Report Analyses and Calculations
How does JMP Clinical handle date and time information?
Times and dates are an integral part of the data generated in all clinical trials. At least one timing variable must be included in all SDTM subject-level domain data sets. JMP Clinical can use time and date variables in a variety of numeric formats; for example, the ISO 8601 standard: YYYY-MM-DDhh:mm:ss, shown here:
| • | YYYY is the four-digit year, |
| • | MM is the two-digit month (values rage from 01-12), |
| • | DD is the two-digit day (values range from 01-31), |
| • | hh is the two-digit hour (values range from 00-23) |
| • | mm is the two-digit minutes (values range from 00-59), and |
| • | ss is the two-digit seconds (values range from 00-59). |
Additional, allowable characters include
| • | T, which indicates that time information is included (omitted if no time component is included), |
| • | -, which either separates the date elements or can be used to indicate missing date components |
| • | :, which separates time elements, |
| • | /, which can be used to separate the date components from the time components, and |
| • | P, which serves as a duration indicator and precedes the date/time components representing the duration of an event or intervention. |
Dates and times can be and are expressed as complete dates/times, partial date/times, or incomplete date/times. JMP Clinical recognizes each of these elements and handles partial or incomplete dates/times as described in the separate FAQ: How does JMP Clinical handle partial or incomplete date and time information?
How does JMP Clinical handle partial or incomplete date and time information?
When date/time values are either partial or incomplete, JMP Clinical enables you to choose to invoke either a “first moment” or 'last moment' rule to handle partial dates. In these cases, when a date/time component is not included in the ISO 8601 value, that value is assumed to be the first or last possible value, depending on your selection.
If the imputation rule for partial date/time values is "First":
| • | if the time is missing, JMP Clinical imputes 00 missing hours, minutes, and seconds. |
| • | if the day is missing, JMP Clinical imputes the first day of the month. |
| • | if both the day and the month are missing, JMP Clinical imputes the first day of the first month (01-Jan). |
If imputation method for partial date/time values is "Last":
| • | if the time is missing, JMP Clinical imputes 23 hours, 59 minutes, and 59 seconds. |
| • | if the day is missing, JMP Clinical imputes the last day of that month. |
| • | if both the day and the month are both missing, JMP Clinical imputes 31 for day and 12 for month (31-Dec). |
For example, you have chosen to use the first rule and the date value for an event is listed as “2013-12”. In this case, JMP Clinical assumes that the event occurred at 12:00 AM on the first day of December 2013. and assigns a time value of “00:00" and a day value of "01". The resulting sort order of partial dates using the first-moment rule is the same as that of the ISO 8601 strings themselves.
How does JMP Clinical derive date and time information?
Conversion of Date/Time variables from Categorical to Numeric format
Date and time variables in CDISC-formatted data typically follow the ISO 8601 standard: YYYY-MM-DDhh:mm:ss and, thus, are categorical variables. To derive study day values, JMP Clinical must first convert those variables into numeric variables. It does this by first calculating the number of seconds that have occurred between the date and time listed for an event/finding/intervention and a reference date of 12:00 AM on January 1, 1904 and replacing the categorical value with the numeric elapsed seconds value. JMP Clinical automatically does this conversion. While this numeric date may not be displayed in your data table, rest assured that it has been calculated and stored behind the scenes.
Calculating Study Day
JMP Clinical derives the study day from the date/time information as follows:
| • | If the event/finding/intervention occurred after the anchor date (either the treatment start date or reference start date, usually RFSTDTC), JMP Clinical subtracts the reference date from the date of the event/finding/intervention and then adds 1. For example, if treatment started on March 10th and a finding was made on Mar 20th, the study day of the finding would be (20 - 10) + 1 = 11. |
| • | If the event/finding/intervention occurred before the anchor date (during prescreening, for example), JMP Clinical still subtracts the reference date from the date of the event/finding/intervention but does not add 1. For example, if treatment started on March 10th and a finding was made on Mar 5th, the study day of the finding would be (5 - 10) = -5. |
Note: You can opt to use either the reference start date or the treatment start date as the anchor date. See Set Study Preferences for more information.
How does JMP Clinical identify domains included in a study?
Domains are evaluated for SDTM folder and ADaM folder. Domains from SDTM folder are named using the 2-letter code XX, where XX. can be any two letters. For example, the domain containing adverse event data is AE (the data set name).
Domains from ADAM always contain AD as the first two letters of the domain name. ADSL is constant while other letters following the AD identify the specific domain. For example, ADAE is the ADaM domain AE.
Domains are classified as findings if XXTESTCD is present, interventions if XXTRT is present, or events if XXTERM is present. If domain type cannot be identified for a given folder, the domain is ignored.
In addition, Basic Data Structure (BDS) is supported for ADAM. If PARAMCD and either AVAL or AVALC are present, the domain is considered a findings domain. If other variables are present as above and the domain type cannot be identified, it is ignored. This is true even if XXTESTCD is present since it would not be clear whether to transform ADaM variables or use SDTM variables for the domain.
Once domains are selected for each folder, each domain is represented by one data set. If a domain is present in both ADAM and SDTM, the ADAM domain takes precedence.
The one exception to ADaM's precedence occurs when the DM domain from SDTM contains variables not present in ADSL. In this case, DM is merged with ADSL. Variables that are unique to one data set or the other are all included in the merged data set. However, for variables common to both ADSL and DM, the ADSL variable takes precedence and is included in the merged data set, the DM version is discarded. if the study includes SUPPDM, then DM and SUPPDM are merged prior to the merge with ADSL.
Supplemental domains (SUPPxx) can be used by JMP Clinical. However, because these domains lack the standard data needed for analysis, SUPPxx domains are recognized only when the main domains are present. For example, SUPPAE is recognized when AE is present but is ignored when AE is not found. SUPP, where all supplemental data is within one data set, is not supported in the CDISC specification and will not be used.
How does JMP Clinical use data contained across split domains?
When you add a study, JMP Clinical examines the specified SDTM and ADaM folders and identifies the domains using the standard two-letter designations.
Split domains are identified by three- or four-letter designations. The first two letters correspond to parent domain with one or two additional characters. Split domains may either be present in the specified SDTM folder with the other domains or may be grouped within a single subfolder located in the SDTM folder. This subfolder must be named split. JMP Clinical adds the study as normal, maintaining the file structure, but subsequently treats the split domains as a single unit during analysis.
Note: The split domains are ignored if the major domain (identified by the two-letter designation) is present.
What are Keys and how does JMP Clinical Identify Keys?
Keys
The CDISC SDTM Implementation Guide defines the following terms:
A Natural Key is one or more variables whose contents uniquely distinguish every record (row) in the data set. For example, each row of the DM domain should represent a different subject. The natural keys in this instance could be Study Identifier (STUDYID) and Unique Subject Identifier (USUBJID).
A Surrogate Key is an artificially established single-variable identifier that uniquely identifies rows. This could include any of the xxSEQ variables. For example, if the vital signs data set contained 200 records, the VSSEQ variable could be numbered 1 to 200 to uniquely identify the rows.
Alternatively, xxSEQ can be made part of a natural key so that xxSEQ can count from 1 to ni, where ni is the total number of records for a subject. Here the keys would be STUDYID, USUBJID and xxSEQ.
How does JMP Clinical Identify Keys?
JMP Clinical looks for keys using the following sequence:
| 1 | Text Files |
JMP Clinical looks for presence of xx.txt files (for SDTM) or ADxx.txt files (for ADaM) in keys subfolders located in either the SDTM or ADaM folders where the domains are stored. For each domain (DM, CO, SE, SV, the findings domains, the events domains, and the interventions domains) and ADSL, the JMP Clinical chooses either the xx.txt file (where xx is the domain name) or the ADxx.txt file depending on the library used for each domain. Keys are derived from that text file.
For example, a VS.txt file located in the SDTM folder contains the following five rows:
USUBJID
VSTESTCD
VISITNUM
VSDTC
VSGRPID
The three variables defined in this text file serve as keys for the analysis.
If keys are not defined, JMP Clinical proceeds to step 3.
Note: Keys files need to present during updates to enable users to change keys during snapshots if needed to account for non-uniqueness of rows based on a past set of keys.
If keys are still not defined, JMP Clinical proceeds to step 4.
Note: This option is not available for SAS Transport files.
| 2 | Derive |
Keys are derived based on the suggested variables above.
How does JMP Clinical determine whether an Event Is a Treatment Emergent Adverse Event?
JMP Clinical uses a JMP Event Type function to determine whether records might be:
| • | Treatment emergent. These are data that begin on or after the first dose of any study drug. |
| • | Pre-treatment. These are data that begin prior to the first dose of any study drug. |
| • | On Treatment. Those are events that begin on or after the first dose of any study drug until the last date of dosing plus the offset1 for end of dosing |
| • | Off-treatment follow-up. These are events that begin after the last date of dosing plus the offset for the end of dosing |
This function can be applied to event domains (including AE and CE), intervention domains (including CM) and findings domains (including VS) and supplemental domains (including SUPPAE).
Algorithm
The function used to calculate TEAEs incorporates the following:
| • | Dates with partial times are imputed to the earliest time, 00 seconds, 00 minutes, or 00 hours. |
| • | Partial dates impute to earliest day (1st), and earliest month (Jan). Dates that are completely missing are not imputed. Observations recorded in MH are assumed to have occurred prior to initial dosing. |
| • | If either ADSL.TRTEMFL or xx.xxTRTEM are present with a value of either Yes or Y, the event is considered as treatment emergent. |
| • | The start dates for dosing are determined from ADSL.TRTSDTM. If ADSL.TRTSDTM is not present, ADSL.TRTSDT is used. If ADSL.TRTSDT is not present, DM.RFXSTDTC is used. If DM.RFXSTDTC is not present, the earliest date in EX.EXSTDTC is used. If EX.EXSTDTC is not present, DM.RFSTDTC is used. |
| • | The end dates for dosing are determined from ADSL.TRTEDTM. If ADSL.TRTEDTM is not present, ADSL.TRTEDT is used. If ADSL.TRTEDT is not present, DM.RFXENDTC is used. If DM.RFXENDTC is not present, the latest date in EX.EXENDTC is used. If EX.EXENDTC is not present, DM.RFENDTC is used. |
| • | If neither the date of the event, intervention or finding nor the dosing start date has a time component, treatment emergence status is based solely on the date. |
| • | For AE, CM, and CE, if the date listed is on or after the dosing start date, the event is considered treatment emergent. |
| • | All non-treatment emergent events are considered pre-treatment events. |
| • | Note: All events are considered pre-treatment for those subjects not on treatment. |
| • | For conservativeness, AE and CM are considered treatment emergent and on-treatment when equal to dose datetime or when the date is missing. |
| • | For on-treatment events: |
| • | When neither the date of event, nor the intervention of finding nor the dosing end date has a time component, comparisons are based solely on date. |
| • | For those cases in which the start date is known but the end date is missing, comparisons are based solely on the date and the event is considered on-treatment when the date of the event occurs after the start date. |
| • | For those cases in which both the start date and the end date are known, comparisons are based solely on the date and the event is considered on-treatment when the date of the event occurs between the start date and end date (plus the offset). |
| • | For off-treatment follow-up events: |
| • | When neither the date of event, nor the intervention of finding nor the dosing end date has a time component, comparisons are based solely on date. |
| • | For those cases in which both the start date and the end date are known, the event is considered off-treatment follow-up when the date of the event occurs after the end date (plus the offset). |
How does JMP Clinical handle non-unique Findings test names?
When running Findings reports, JMP Clinical looks for and appends the values from either xxPOS or xxSPEC to the test names in xxTESTCD and xxTEST. This enables you to analyze findings data when multiple findings test names are identical across the variables: xxTESTCD, xxPOS, and xxSPEC.
If test name values are still not unique across categories of xxCAT or xxSCAT (if they exist) after appending the prior variables, a numeric index is appended to non-unique tests so that reports can still be run and tests are not inappropriately combined.
How does JMP Clinical derive reference range and baseline reference range indicators?
If ADaM is present, it takes precedence over SDTM and JMP Clinical reads the Reference Range Indicators and Baseline Reference Range Indicators from the Analysis Range Indicator Variable (ANRIND) and the Baseline Range Indicator Variable (BNRIND), respectively. If either of these are absent, then they can be constructed from the Analysis Normal Range Lower Limit (ANRLO) and the Analysis Normal Range Upper Limit (ANRHI)
If ADaM is not present, SDTM variables are used. In this case JMP Clinical uses the values of the Reference Range Indicator Variable (LBNRIND) for Reference Range Indicators and Baseline Reference Range Indicators. If the Reference Range Indicator Variable (LBNRIND) variable is absent, then the Reference Range Lower Limit-Standard Units (LBSTNRLO) and the Reference Range Upper Limit-Standard Units (LBSTNRHI) can be used to construct the values of the Reference Range Indicator Variable and Baseline Range Indicator Variable.
How does JMP Clinical handle supplemental domains?
The Merge supplemental domain option is checked by default in reports where the domain data format remains intact and supplemental can be merged into the output data set. Additionally, reports involving adverse events will have merge supplemental checked by default due to common use of SUPPAE to store treatment emergent flags. All other reports that process or summarize data have the Merge supplemental domain option off by default to improve efficiency and processing of the reports.
If you wish to use information available in a supplemental domain in filtering the data set with custom WHERE statements, then the Merge supplemental domain option should be checked.
How does JMP Clinical Identify Records across Snapshots?
Keys give us insight into the uniqueness of a record or data set row. The CDISC SDTM Implementation Guide defines the following terms:
| • | Natural Keys are one or more variables whose contents uniquely distinguish every record (row) in the data set. For example, each row of the DM domain should represent a different subject. The natural keys in this instance could be Study Identifier (STUDYID) and Unique Subject Identifier (USUBJID). |
| • | A Surrogate Key is an artificially established single-variable identifier that uniquely identifies rows. This could include any of the xxSEQ variables. For example, if the vital signs data set contained 200 records, the VSSEQ variable could be numbered 1 to 200 to uniquely identify the rows. |
Alternatively, xxSEQ can be made part of a natural key so that xxSEQ can count from 1 to ni, where ni is the total number of records for a subject. Here the keys would be STUDYID, USUBJID, and xxSEQ.
So why is this important? Well, in order to examine a record (row) for differences between two snapshots, there needs to be a way to link these two versions of the record together. This is where the keys come in. Otherwise, JMP Clinical has no way to know which records to match together. Further, in order to save or access notes for a particular record, there needs to be a way to file the note away so that it is accessible later when returning to the record. Again, this is where the keys come in.
If xxSEQ surrogate key is available in the data, it will be used as a first choice to uniquely identify records. The xxSEQ variable must be carefully maintained so that the number never changes for a particular record. For example, suppose CM domain contains two records:
|
CMSEQ |
CMTRT |
CMSTDTC |
|
1 |
ASPIRIN |
03-20-1974 |
|
2 |
IBUPROFEN |
and is updated through query with a new medication that actually falls between the first two based on date:
|
CMSEQ |
CMTRT |
CMSTDTC |
|
1 |
ASPIRIN |
03-20-1974 |
|
2 |
IBUPROFEN |
|
|
3 |
VITAMIN C |
03-24-1974 |
It is important that any new records are tacked at the end (and to continue the sequence of CMSEQ). Alternatively, if a record is deleted:
|
CMSEQ |
CMTRT |
CMSTDTC |
|
2 |
IBUPROFEN |
|
|
3 |
VITAMIN C |
03-24-1974 |
The sequence number must be kept consistent (in other words, 1 can never be used again).
The following tables show the keys used by JMP Clinical to uniquely identify records. The first choice is marked with (1), the second choice is marked with (2), etc. If any of the variables from the (1) choice is missing, then JMP Clinical will use the (2) choice, and if any of the variables from the (2) choice is missing, then JMP Clinical will use the (3) choice, and so on. The (1) choice usually contains xxSEQ, the (2) choice is based on the SDTM Implementation Guide.
|
Class |
SDTM Domain |
Keys |
| Special Purpose | DM | (1) STUDYID, USUBJID |
| CO |
(1) STUDYID, USUBJID, COSEQ (2) STUDYID, USUBJID, IDVAR, COREF, CODTC |
|
| SE |
(1) STUDYID, USUBJID, SESEQ (2) STUDYID, USUBJID, ETCD, SESTDTC |
|
| SM |
(1) STUDYID, USUBJID, SMSEQ (2) STUDYID, USUBJID, MIDS |
|
| SV | (1) STUDYID, USUBJID, VISITNUM | |
| Interventions | CM, EX, EC, AG, ML, PR, SU, etc. |
(1) STUDYID, USUBJID, xxSEQ (2) STUDYID, USUBJID, xxTRT, xxSTDTC |
| Events | AE, DS |
(1) STUDYID, USUBJID, xxSEQ (2) STUDYID, USUBJID, xxDECOD, xxSTDTC (3) STUDYID, USUBJID, xxTERM, xxSTDTC |
| MH |
(1) STUDYID, USUBJID, xxSEQ (2) STUDYID, USUBJID, xxDECOD (3) STUDYID, USUBJID, xxTERM |
|
| CE, DV, HO |
(1) STUDYID, USUBJID, xxSEQ (2) STUDYID, USUBJID, xxTERM, xxSTDTC |
|
| All other domains |
(1) STUDYID, USUBJID, xxSEQ (2) STUDYID, USUBJID, xxDECOD, xxSTDTC (3) STUDYID, USUBJID, xxTERM, xxSTDTC |
|
| Findings | CV, EG, FT, MB, MS, PC, RE, VS |
(1) STUDYID, USUBJID, xxSEQ (2) STUDYID, USUBJID, xxTESTCD, VISITNUM, xxTPTREF, xxTPTNUM |
| IS, SS, PE, RP |
(1) STUDYID, USUBJID, xxSEQ (2) STUDYID, USUBJID, xxTESTCD, VISITNUM |
|
| DA, DD |
(1) STUDYID, USUBJID, xxSEQ (2) STUDYID, USUBJID, xxTESTCD, xxDTC |
|
| IE, SC |
(1) STUDYID, USUBJID, xxSEQ (2) STUDYID, USUBJID, xxTESTCD |
|
| FA, SR |
(1) STUDYID, USUBJID, xxSEQ (2) STUDYID, USUBJID, xxTESTCD, xxOBJ, VISITNUM, xxTPTREF, xxTPTNUM |
|
| MK, MO |
(1) STUDYID, USUBJID, xxSEQ (2) STUDYID, USUBJID, VISITNUM, xxTESTCD, xxLOC, xxLAT |
|
| UR |
(1) STUDYID, USUBJID, xxSEQ (2) STUDYID, USUBJID, VISITNUM, xxTESTCD, xxLOC, xxLAT, xxDIR |
|
| NV |
(1) STUDYID, USUBJID, xxSEQ (2) STUDYID, USUBJID, VISITNUM, xxTPTNUM, xxLOC, xxTESTCD |
|
| OE |
(1) STUDYID, USUBJID, xxSEQ (2) STUDYID, USUBJID, VISITNUM, FOCID, xxTESTCD, xxTSTDTL, xxMETHOD, xxLOC, xxLAT, xxDIR, VISITNUM, xxDTC, xxTPTREF, xxTPTNUM, xxREPNUM |
|
| MI |
(1) STUDYID, USUBJID, xxSEQ (2) STUDYID, USUBJID, xxSPEC, xxTESTCD |
|
| LB |
(1) STUDYID, USUBJID, xxSEQ (2) STUDYID, USUBJID, xxTESTCD, xxSPEC, VISITNUM, xxTPTREF, xxTPTNUM |
|
| QS |
(1) STUDYID, USUBJID, xxSEQ (2) STUDYID, USUBJID, xxCAT, xxSCAT, VISITNUM, xxTESTCD |
|
| PP |
(1) STUDYID, USUBJID, xxSEQ (2) STUDYID, USUBJID, xxTESTCD, xxCAT, VISITNUM, xxTPTREF |
|
| RS |
(1) STUDYID, USUBJID, xxSEQ (2) STUDYID, USUBJID, xxTESTCD, VISITNUM, xxTPTREF, xxTPTNUM, xxEVAL, xxEVALID |
|
| TR |
(1) STUDYID, USUBJID, xxSEQ (2) STUDYID, USUBJID, xxTESTCD, xxEVALID, VISITNUM |
|
| TU |
(1) STUDYID, USUBJID, xxSEQ (2) STUDYID, USUBJID, xxEVALID, xxLINKID |
|
| All other domains | (1) STUDYID, USUBJID, xxSEQ | |
| Trial Design | TA | (1) STUDYID, ARMCD, TAETORD |
| TD | (1) STUDYID, TDORDER | |
| TE | (1) STUDYID, ETCD | |
| TI | (1) STUDYID, IETESTCD | |
| TM | (1) STUDYID, MIDSTYPE | |
| TS | (1) STUDYID, TSPARMCD, TSSEQ | |
| TV | (1) STUDYID, ARM, VISIT | |
| Relationships | RELREC | (1) STUDYID, RDOMAIN, USUBJID |
| RELSUB | (1) STUDYID, USUBJID, RSUBJID, SREL | |
| Supplemental Qualifiers | SUPPDM | (1) STUDYID, RDOMAIN, USUBJID |
| All other SUPP domains | (1) STUDYID, RDOMAIN, USUBJID, IDVAR, IDVARVAL |
|
Class |
ADaM Domain |
Keys |
| Special Purpose | ADSL | (1) STUDYID, USUBJID |
| ADCO |
(1) STUDYID, USUBJID, COSEQ (2) STUDYID, USUBJID, IDVAR, COREF, CODTC |
|
| ADSE |
(1) STUDYID, USUBJID, SESEQ (2) STUDYID, USUBJID, ETCD, SESTDTC |
|
| ADSM |
(1) STUDYID, USUBJID, SMSEQ (2) STUDYID, USUBJID, MIDS |
|
| ADSV | (1) STUDYID, USUBJID, AVISITN | |
| Interventions | ADCM, ADEX, ADEC, ADAG, ADML, ADPR, ADSU, etc. |
(1) STUDYID, USUBJID, xxSEQ (2) STUDYID, USUBJID, ASEQ (3) STUDYID, USUBJID, xxTRT, xxSTDTC (4) STUDYID, USUBJID, xxTRT, ASTDTM (5) STUDYID, USUBJID, xxTRT, ASTDT (6) STUDYID, USUBJID, xxTRT, ADT |
| ADAE, ADDS |
(1) STUDYID, USUBJID, xxSEQ (2) STUDYID, USUBJID, ASEQ (3) STUDYID, USUBJID, xxDECOD, xxSTDTC (4) STUDYID, USUBJID, xxDECOD, ASTDT (5) STUDYID, USUBJID, xxTERM, xxSTDTC (6) STUDYID, USUBJID, xxTERM, ASTDT |
|
| ADMH |
(1) STUDYID, USUBJID, xxSEQ (2) STUDYID, USUBJID, ASEQ (3) STUDYID, USUBJID, xxDECOD (4) STUDYID, USUBJID, xxTERM |
|
| ADCE, ADDV, ADHO |
(1) STUDYID, USUBJID, xxSEQ (2) STUDYID, USUBJID, ASEQ (3) STUDYID, USUBJID, xxTERM, xxSTDTC (4) STUDYID, USUBJID, xxTERM, ASTDT |
|
| All other domains |
(1) STUDYID, USUBJID, xxSEQ (2) STUDYID, USUBJID, ASEQ (3) STUDYID, USUBJID, xxDECOD, xxSTDTC (4) STUDYID, USUBJID, xxDECOD, ASTDT (5) STUDYID, USUBJID, xxTERM, xxSTDTC (6) STUDYID, USUBJID, xxTERM, ASTDT |
|
| Findings | All Domains |
(1) STUDYID, USUBJID, xxSEQ (2) STUDYID, USUBJID, ASEQ (3) STUDYID, USUBJID, PARAMCD, AVISITN, ATPTN |
See the SDTM Implementation Guide for more information.
How does JMP Clinical perform Crossover Analysis?
Refer to Crossover Analyses.
How are disposition events used in different JMP Clinical reports?
JMP Clinical reports reports use and count disposition events in specific ways that differ between reports. Standard Safety Reports and DSUR/PSUR Report count subjects in only the last disposition event they experience, whereas DS Distribution (see Events Distribution) counts a subject once in each disposition event they experience. Mortality Time to Event only shows the disposition event only for censored subjects, so if a subject died they are counted under Death and not for any other disposition event.
How does JMP Clinical calculate the False Discovery Rate (FDR)?
With J treatment comparisons of ordered (smallest to largest) p-values p(j), the FDR p-value (Benjamini and Hochberg, 19952) for the jth hypothesis is:
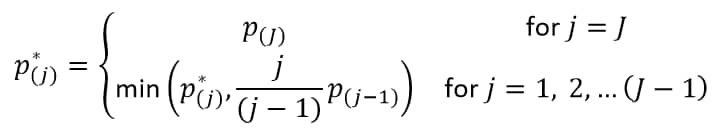
The reference line is drawn at the -log10 transformation of the maximum raw p-value where the corresponding FDR p-value is less than or equal to .
How are risk thresholds defined?
A risk threshold data set is used to 1) define the risk levels for individual variables for RBM analyses 2) specify the contribution of each variable to overall indicators of site risk.
In general, risk thresholds work the same for individual indicators as they do for Overall Indicators and can be defined as described below:
For the ith site or country and the jth risk indicator,
| • | Moderate risk (yellow) occurs when |
| • | Severe risk (red) occurs when |
where is the mean, median or user-supplied center value. The quantity
equals
,
, and
, for Direction of Risk Signals equal to B, U, and L, respectively, and
is the value for the it h site or country and the jth risk indicator.
It is acceptable to specify both yellow and red risk thresholds, one or no risk thresholds. When specifying only a moderate threshold, the Red Percent of Center is left missing in the risk threshold data set so that moderate risk is considered. In instances where values do not meet the criteria for moderate or severe risk, the risk is considered mild (green). Note that for risk thresholds defined using the above criteria, no threshold colors are determined in instances where the mean, median or center value is calculated or set to zero.
Risk thresholds can also be defined based solely only the magnitudes of the values observed. In this case,
| • | either moderate risk (yellow) occurs when |
| • | severe risk (red) occurs when |
where the quantity is defined as described above.
In this case, it is acceptable to specify both thresholds, one threshold, or no risk thresholds at all. When specifying only a moderate threshold, the Red Magnitude is left missing in the risk threshold data set so that moderate risk is . In cases where neither moderate nor severe risk applies, the risk is considered mild (green).
There are five overall risk indicators. These are either weighted averages or combinations of the individual risk indicators for which at least one risk threshold is defined, where both the Weight for Overall Risk Indicator and the standard deviation of the indicator > 0.
The first, or Overall Risk Indicator, incorporates all of the variables meeting these criteria into a single measure that signifies the overall risk and performance of a clinical site. This indicator is generated only when the Weight for Overall Risk Indicator exceeds 0 for at least one of the available risk indicators exhibiting variability. If none of the individual indicators have a Weight for Overall Risk Indicator > 0, then the corresponding Overall Risk Indicator is not generated.
Each of the other four overall indicators - Enrollment Metrics, Disposition, Adverse Events, and Manually Entered - combines subsets of the risk indicators based on Category in the risk weight data set. By default, Category matches how variables are grouped in Risk-Based Monitoring, with Manually Entered applied to all user-supplied risk indicators from Update Study Risk Data Set. If no indicators have a Weight for Overall Risk Indicator > 0 for a given category, then the corresponding overall indicator is not provided.
The Weight for Overall Risk Indicator (wj) can either be missing (in this case, it is assumed to be zero) or greater than or equal to zero. The weights are self-normalizing in that each weight is divided by the sum of all weights for variables contributing to the particular overall indicator. The contribution of each indicator to an overall indicator is based on its weight, center value (either mean, median or user-provided center value, ), standard deviation (
), and direction. In general, the value for an overall indicator for the ith site or country and the jth risk indicator is defined as
, where
,
, or
when Direction equals B,U, or L, respectively. This can be interpreted as larger values imply greater risk. By default, all weights are assumed equal to one in the Default Risk Threshold data set, meaning that each variable contributes equally to each overall indicator.