Cluster Subjects Within Study Sites
This report clusters subjects within study site for the purpose of identifying similar subjects. It constructs a cross-domain data set using as much baseline data as possible (subject to user options). The dataset includes age and the variables from available findings domains (LB, EG, VS, etc.) with less than 5% missing values. Next, it calculates Euclidean distances to compute a distance matrix and performs hierarchical clustering of subjects within each study site. Findings values are averaged by USUBJID, test code, visit number, and time point (if available) if there are multiple measurements for a visit or time point. The goal of this exercise is to identify pairs of subjects with a very small distance. This could be an indication that these subject are slightly modified copies of one another.
Report Results Description
Running this report for Nicardipine using default settings generates the report shown below.
The Cluster Subjects Within Study Sites report shows the results of clustering of the subjects on the basis of different study sites. The results for each grouping are presented in a separate “section”.
Between-Subject Distance Summary
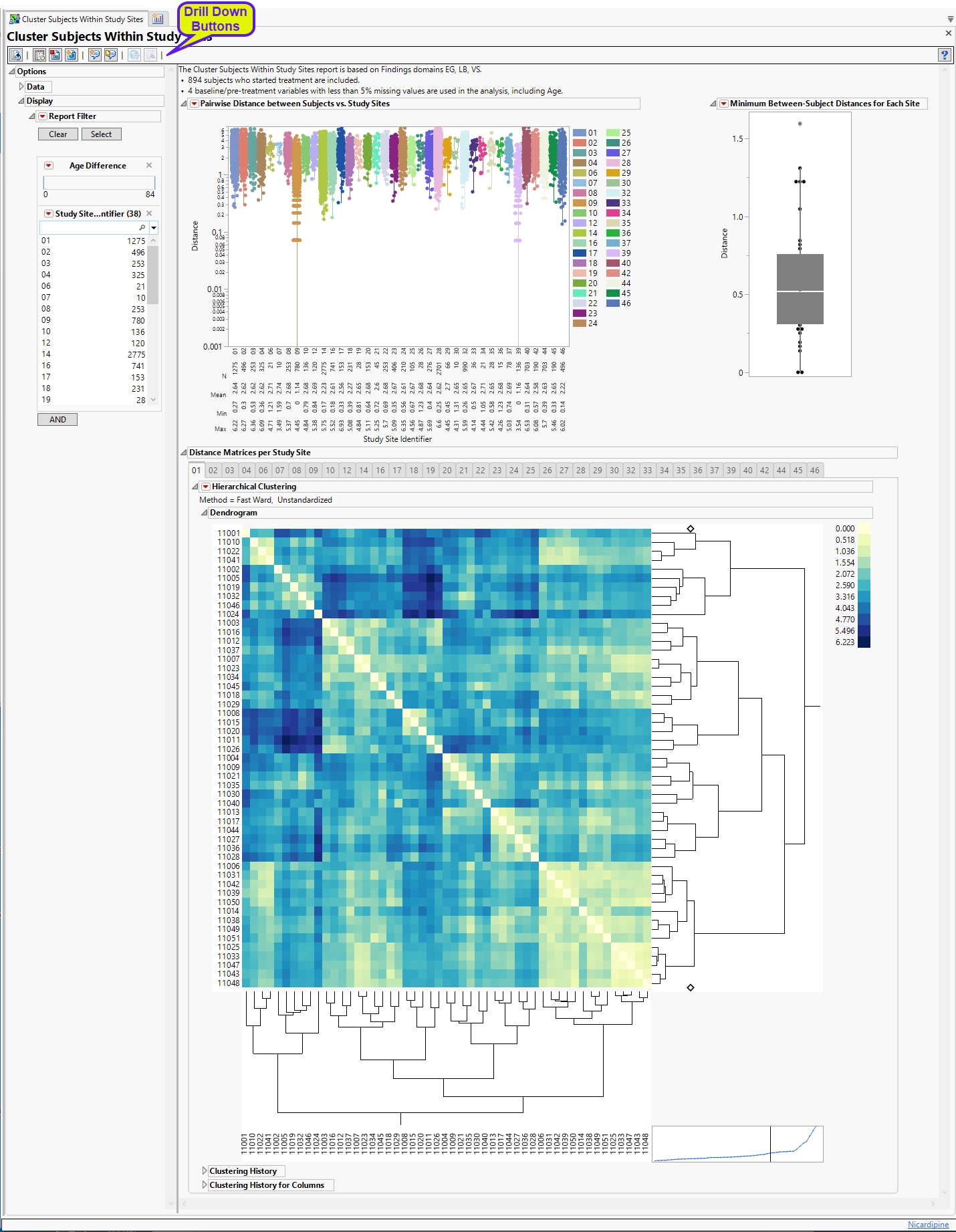
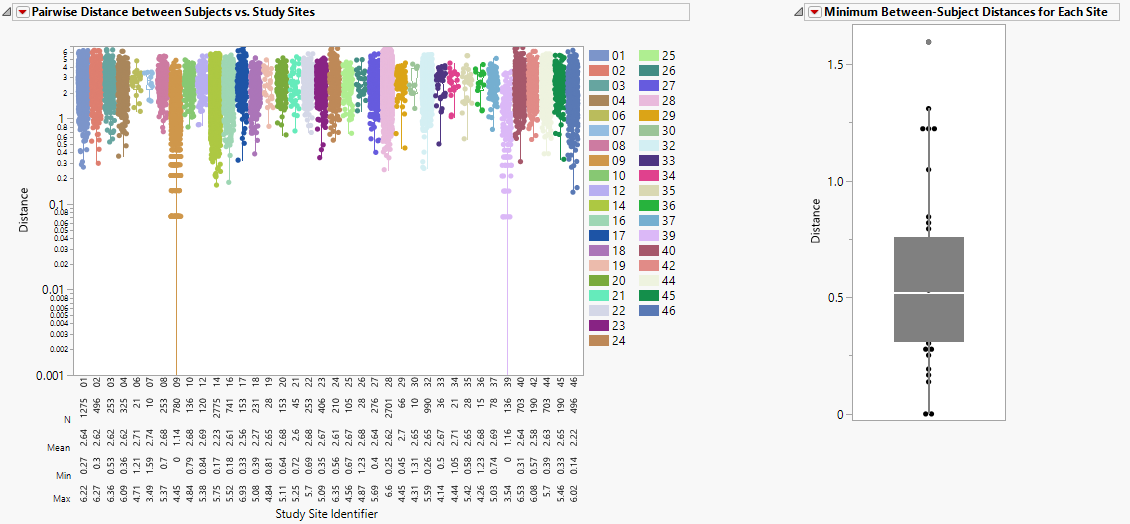
This section presents box plots of the pairwise Euclidian distances between subjects presented by site and Box Plot of the minimum pairwise distance taken from each site.
It contains the following elements:
| • | One set of Box Plots of Pairwise Distances between Subjects vs. Study Sites. |
This figure shows box plots of all pairwise Euclidian distances within each study site. Individual points are overlaid with the boxplot. The Y-axis is set to log-scale by default for the ease of identifying small distance points when the range of distance is large. A summary table is shown below the boxplot presenting the N, Mean, Min, Max values for each study site. Values closer to zero (0) reflect subjects that are very similar to one another, which could indicate that they are slightly modified copies.
| • | One Box Plot of Minimum Between-Subject Distances for Each Site. |
The box plot of minimum pairwise distances provides some idea of the most similar subjects within each site.
Distance Matrices per Study Site
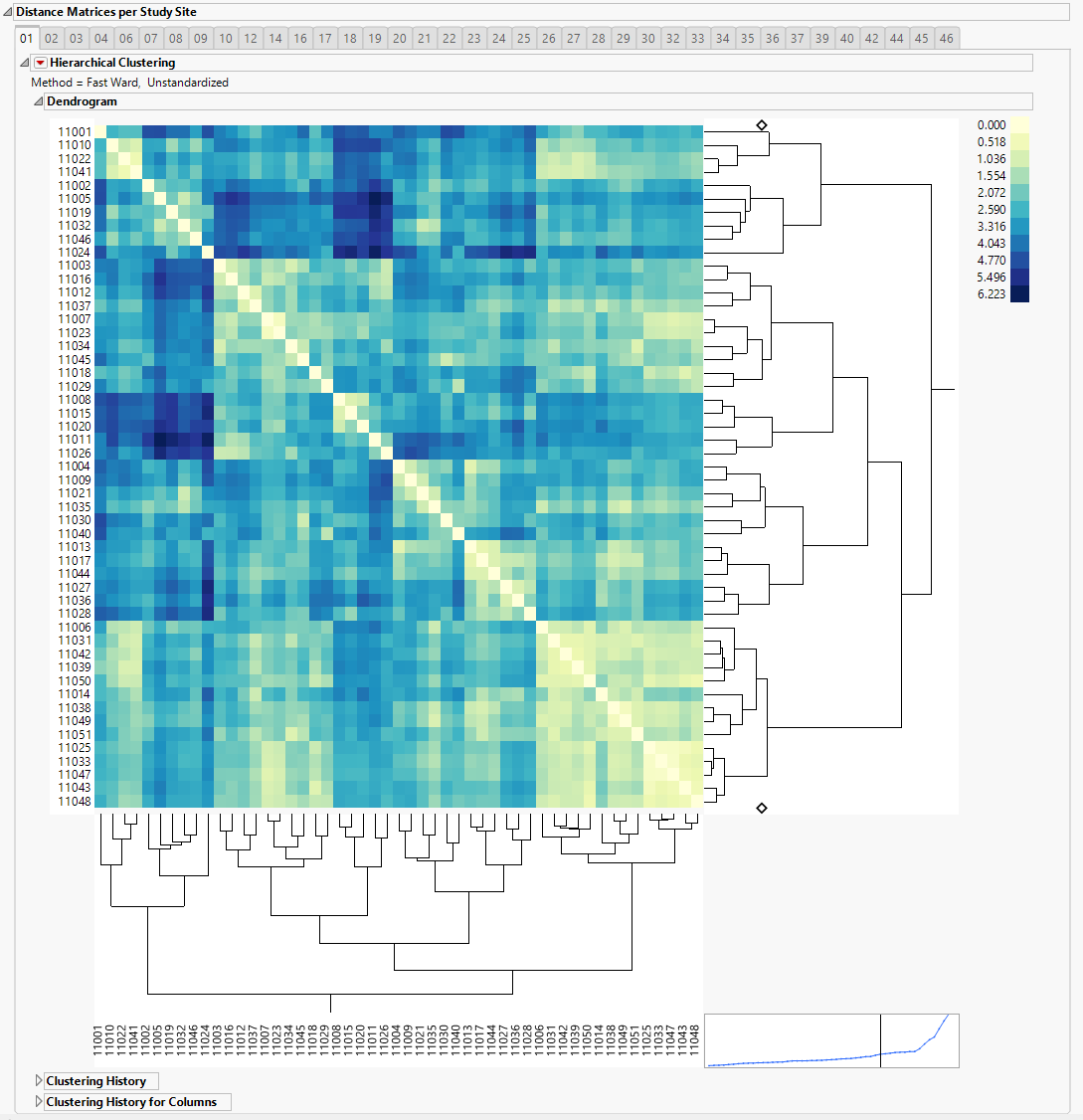
The Distance Matrices per Study Site section is shown below. There is one panel for each study site in the trial. The panels are named after the study sites' names or IDs.
It contains the following elements:
| • | One Hierarchical Clustering display that clusters subjects based on the Euclidean distance. A heatmap from Hierarchical Clustering is shown with subject IDs being rows and columns annotations. The legend shows the pairwise distance values with a minimum of zero. |
Bluer color indicates subjects that are more similar, whereas yellow shows subjects less similar. The clustering dendrogram is presented to the right of the heat map and can show sets of more than two subjects that are similar to one another. All the pairwise distance matrices, along with the subjects' ID , study sites and treatment arms, are stored in separate jmp tables, and can be accessed by clicking the 'Show Tables' button.
Options
Data
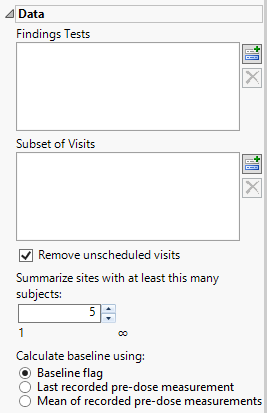
Findings Tests
Use this widget to select Findings Tests for the analysis. The report will autorun and analysis is restricted to the selected tests only.
Subset of Visits
Use the Subset of Visits option to select the visits to be included in the analysis.
Remove unscheduled visits
You might or might not want to include unscheduled visits when you are analyzing findings by visit. Check the Remove unscheduled visits to exclude unscheduled visits.
Summarize sites with at least this many subjects:
The Summarize sites with at least this many subjects: widget enables you to set a minimal threshold for the sites to be analyzed. Only those sites which exceed the specified number of subjects are included. This feature is useful because it enables you to exclude smaller sites, where small differences due to random events are more likely to appear more significant than they truly are. In larger sites, observed differences from expected attendance due to random events are more likely to be significant because any deviations due to random events are less likely to be observed.
Calculate baseline using:
Use the Calculate baseline using: widget to use one of the following: i) baseline flag variable, ii) the last recorded pre-dose measurement, or iii) the mean of all the measurements taken during the baseline time window as the baseline measurement.
Display
Report Filter
These filters enable you to subset and view subjects based on demographic characteristics (Age Difference), study sites, and other criteria. Refer to Data Filter for more information.
General and Drill Down Buttons
Action buttons, provide you with an easy way to drill down into your data. The foclusllowing action buttons are generated by this report:
| • | Click  to rerun the report using default settings. to rerun the report using default settings. |
| • | Click  to view the associated data tables. Refer to Show Tables/View Data for more information. to view the associated data tables. Refer to Show Tables/View Data for more information. |
| • | Click  to generate a standardized pdf- or rtf-formatted report containing the plots and charts of selected sections. to generate a standardized pdf- or rtf-formatted report containing the plots and charts of selected sections. |
| • | Click  to generate a JMP Live report. Refer to Create Live Report for more information. to generate a JMP Live report. Refer to Create Live Report for more information. |
| • | Click  to take notes, and store them in a central location. Refer to Add Notes for more information. to take notes, and store them in a central location. Refer to Add Notes for more information. |
| • | Click  to read user-generated notes. Refer to View Notes for more information. to read user-generated notes. Refer to View Notes for more information. |
| • | Click  to open and view the Review Subject Filter. to open and view the Review Subject Filter. |
| • | Click  to specify Derived Population Flags that enable you to divide the subject population into two distinct groups based on whether they meet very specific criteria. to specify Derived Population Flags that enable you to divide the subject population into two distinct groups based on whether they meet very specific criteria. |
Default Settings
Refer to Set Study Preferences for default Subject Level settings.
Methodology
No testing is performed. Subjects are clustered within each site according to the selected clustering methodology. See statistical details for hierarchical clustering in the JMP documentation.