Variable Gene Selection
Running this process for the PBMC sample setting generates three output data sets accessed from a Results window shown below. Refer to the Variable Gene Selection process description for more information about this process.
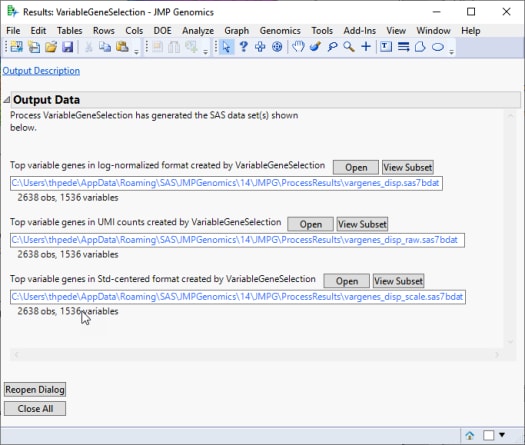
The Results window contains the following pane:
Output Data
This process generates the following output data sets:
| • | A log-Normalized Output Data Set: This output data set includes selected variable genes with the log-normalized values replacing the raw count values. This data set can be used as input for a wide variety of analytical processes include Basic RNA-Seq Workflow and Basic Single Cell RNA-Seq Workflow. |
| • | Original Count Data Set: The output data set (identified by the appended _raw suffix) includes selected variable genes with the original UMI count data. This data set can be directly used for data visualizations. It can also be used as input for a wide variety of analytical processes after appropriate data normalization procedures. |
| • | Standardized Data Set: The output data set (identified by the appended _scale suffix) includes selected variable genes with the standard deviation-standardized values replacing the raw count values. This data set can be used as input for PCA, clustering, and other multivariate analyses. |
Note: You should decide which data set should be used based on the type of analysis or visualization you want to do. The Basic Single Cell RNA-Seq Workflow uses a log-normalized data set by default.
General
| • | Click to reopen the completed process dialog used to generate this output. |
| • | Click to close all graphics windows and underlying data sets associated with the output. |