Launch the Marker Simulation Platform
To simulate a genetic cross from your experimental data, launch the Marker Simulation platform by selecting Analyze > Genetics > Marker Simulation.
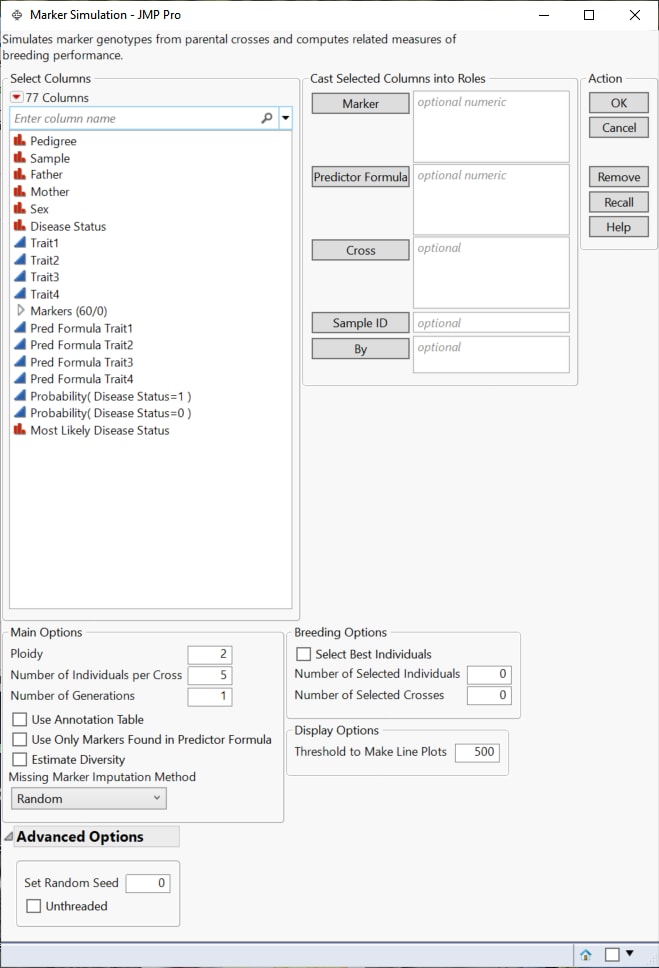
Figure 4.3 Marker Simulation Launch Window
Marker
Select desired marker columns and click Marker to specify the markers that you want to analyze.
Predictor Formula
Use this option to specify columns containing the predictor formulas. These formulas are developed on historical data where an event has been measured or inferred. They are generated using one or more predictive modeling processes using predictive platforms in JMP, for example, Fit Model, Response Screening, XGBoost, etc. The predictive models are then applied to new data for which the attributes are known, but the event has not yet occurred.See Generating Predictor Formulas for Marker Simulation for details.
Note: Any trait column lacking a corresponding Predictor Formula column is ignored during the simulation.
Cross
Use this option to specify the column to be used to differentiate the parents in the crosses. Specifying Sex, for example, directs the platform to cross parents of different sex (male with female) only.
Sample ID
Use this option to specify one or more variables whose values that can, either singly or in combination, provide a unique identifier for each row.
By
Produces a separate report for each level of the By variable. If more than one By variable is assigned, a separate report is produced for each possible combination of the levels of the By variables.
Ploidy
Enables you to specify the ploidy level of the experimental organism under investigation. Note: This must be an even number
Number of Individuals per Cross
Enables you to specify the number of replicates.
Number of Generations
Specifies the number of generations.
Use Annotation Table
Enables you to access annotation information contained in a separate data table. After you click OK, a window appears, prompting you to specify the name and location of the annotation table.
Use Only Markers Found in Predictor Formula
Check this box to restrict the simulation to only those markers used to develop the predictor formulas.The algorithms used to generate the predictor formulas typically use some variable section method to select a subset of most significant markers in your data set. You can view the markers used by right-clicking on the column listing the trait predictors and selecting Column Info.
Estimate Diversity
Check this box to calculate estimates of polymorphism, heterozygosity, and allelic diversity and frequency for the progeny of each cross.
Missing Marker Imputation Method
This method does not run when your data is missing marker data. Because of this, any missing data must be imputed. Use this option to specify how the missing values are to be imputed.
– Select HWE Off to impute the missing genotypes with random draws from a multinomial distribution in which the frequency of each genotype class is set to be the observed frequency from the data.
– Select HWE On to impute the missing genotypes with random draws from a multinomial distribution in which the frequency of each genotype class is set to be the expected frequency under the assumption of Hardy-Weinberg Equilibrium.
– Select Random to randomly assign one of the acceptable values (0, 1, 2, ..., K (where K is the ploidy level)).
– Select Specified to impute the missing genotypes with a specified integer between zero and the ploidy number.
Imputation Value
Use this option to specify a value to use in place of any missing genotypes.
To impute with recessive, dominant or heterozygous, first select Specified, then you can enter a number from 0 to ploidy in the Imputation Value box. For diploid organisms, enter 0 for Recessive Homozygous, 1 for Heterozygous, and 2 for Dominant Homozygous Both assuming diploid.
Note: This option is available only when Specified is chosen as the Missing Marker Imputation Method.
Select Best Individuals
Check this box to select only the progeny that meet specified trait criteria, in each generation, for use in the subsequent cross. You must specify the selection criteria for each trait used for the selection. You can specify a lower limit, an upper limit, or a specific target value.
Specify a lower limit to select progeny with a trait value bigger than or equal to this limit to move to next generation. Select an upper limit to select progeny with a trait value lower than or equal to this limit to move to next generation. Specify a target value to select progeny with a trait value equal to this target to move to next generation.
Note: Specification of a target value is done when traits are non-continuous.
You can specify both an upper limit and a lower limit for any given trait to select only the progeny with trait values that fall within the interval formed by the upper and lower limit. Specification of a target value together with either an upper or a lower limit is not valid.
The final selection criterion is the intersection of all criteria specified for the traits. For example, if Spec Limits are such that, L1<= Trait1, L2 <= Trait2 <= U2, and Trait3 == T3, then the selection criterion is constructed to be L1<= Trait1 and L2 <= Trait2 <= U2 and Trait3 == T3. Any progeny that satisfies this criterion will be selected to the next generation.
See Specifying Trait Selection Criteria for Marker Simulation for details about how to specify criteria for selecting progeny.
Note: This option is ignored unless Spec Limits have been specified for at least one of the Predictor Formula columns.
Number of Selected Individuals
This option enables you to specify an upper limit of progeny meeting the trait selection criteria, in each generation, to use as parents in the subsequent cross. This limit is applied repeatedly for each subsequent generation.
Number of Selected Crosses
This option enables you to specify an upper limit on the number of crosses meeting the trait selection criteria. Progeny from the previous cross are assessed for the selection criteria and this limit is then applied, if needed, to the subsequent cross. This limit is applied repeatedly for each subsequent generation.
Threshold to Make Line Plots
Generating line plots representing multi-generational crosses require substantial computer resources; trying to generate too many can overwhelm your computer’s resources. Use this option to set an upper limit to the number of crosses used for generating the line plots. Should the number of crosses made exceed the specified value, JMP does not attempt to generate these plots.
Set Random Seed
Use this option to specify a nonnegative integer to start the random number stream. Different values produce different outcomes of the algorithm.
Unthreaded
Suppresses multi-threading. Deselect this option for improved computational speed.
Required Data Format for the Marker Simulation Platform
Most of the processes in JMP assume that the input table has a particular data structure. JMP distinguishes between tall and wide data sets. A tall data table has samples as columns and molecular entity (for example, marker, gene, clone, protein, or metabolite) as rows, whereas a wide data table is the transpose of the tall data table, having the samples as rows and molecular entity as columns.
When specifying the input data set for a process, it is important to know the required form. Marker Simulation requires a wide data table. The Transpose platform under the Tables menu enables you to transform your data tables between tall and wide forms.
Marker data must be encoded in the one-column, numeric format. Typically, in this format, diploid individuals homozygous for the least common, or minor allele, are represented in the table by a 2, whereas the heterozygotes are represented by a 1. Homozygotes for the most common allele are represented by a 0.