Running a Process
To run a process, select the process from the Starter window or menu, specify the parameters on all tabbed panes in the process dialogs, and then click . The following example, which invokes the ArrayTrack Input Engine, illustrates a typical JMP Genomics process.
| 8 | Select Genomics > Import > Other Expression > ArrayTrack. The dialog shown below opens: |
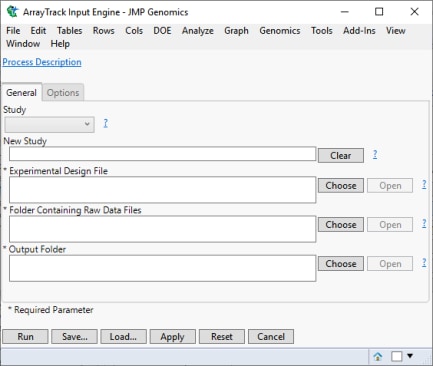
Each dialog has three main sections: a description box, one or more tabbed parameter panes, and functional buttons (illustrated below). The description box on the top of the dialog describes the purpose of the process. The tabbed panes are the main area to specify input parameters.
The eight functional buttons, common to all of the JMP Genomics dialogs, are described below.
|
Functional Button |
Used to: |
|
|
Run the process using the specified parameters |
|
|
Save the specified parameters |
|
|
Load selected, saved parameters into the dialog1 |
|
|
Apply the specified parameters as default settings to all relevant JMP Life Sciences dialogs |
|
|
Save the specified parameters as the default setting. |
|
|
Clear all the parameter settings and return the dialog to its default state |
|
|
Opens this online Help System |
|
|
Cancel the process and close the dialog |
Use these buttons to load, save, or clear specified parameters, run the process using the specified parameters, or apply those parameters to other JMP Life Sciences processes.
There is a defined order to the specification of some parameters. Such parameters are disabled and grayed out until their dependency requirements are fulfilled. Many processes contain multiple tabbed panes with numerous optional parameters. As you develop expertise with particular processes, be sure to investigate the often rich collection of parameters available.
| 8 | Click 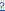 to the right of any parameter entry field to obtain help about its specification. to the right of any parameter entry field to obtain help about its specification. |
The General tab for each dialog typically contains the most important parameters for the process. For example, most processes require specific types of input files or data sets and an output folder. For our example, we want to open the AT_exp2.txt file. This Experimental Design File (EDF), which contains information about the experiment, is needed to import raw data into JMP Genomics.
| 8 | Click . |
| 8 | Click on the AT_exp2.txt file. |
| 8 | Click to select the file (circled below) |

The file is added to the dialog, as shown below.
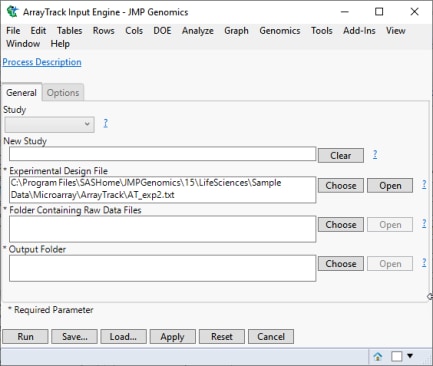
Our next step is to select the folder containing the raw data files.
| 8 | Click (circled below). |

| 8 | Navigate to the Sample Data folder and then to a folder named ArrayTrack by following the path Sample Data > Microarray > ArrayTrack. |
| 8 | Click (circled below) at the bottom of the Browse for Files or Folders window. |
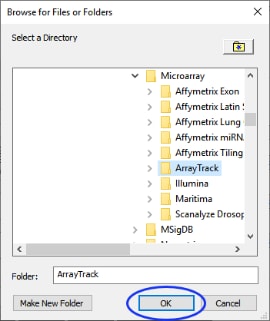
The next step is to choose a folder in which to place and store output. You can choose any folder that you like. For this example, select the ProcessResults folder that came with JMP Genomics.
| 8 | Repeat the selection process to specify the output folder. |
The completed dialog is shown below.
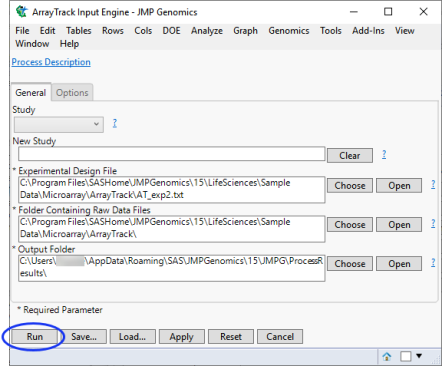
| 8 | Once you have specified the parameters for a process, click to save the parameters for later recall, if needed. |
| 8 | Click to run the process. |
JMP Genomics dialogs generate and run a SAS program each time you click . Depending on the size of your data sets and capacities of your computer, some analyses can take several minutes or, for very large and complex runs, several hours. While a program is running, the message SAS Running Processes window is displayed, as shown below.
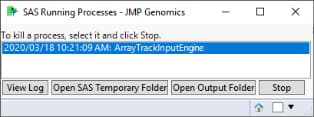
While a process is running, it is a good idea to monitor progress using an application that displays statistics such as CPU, memory, and disk usage, like the Windows Task Manager. This can be informative for troubleshooting a hung process.
You can run only one process at a time. If you attempt to run a second process while another one is running, you are prompted to either disconnect from SAS and stop the current process, to view the current SAS log, or to wait until it completes.
The location of each SAS data set generated by your analysis is listed in a new window (shown below).
