Output Description
LD Block Creation
Running this process for the MoroccoExample sample setting generates the tabbed Results window shown below. Refer to the LD Block Creation process description for more information. Output from the process is organized into tabs. Each tab contains one or more plots, data panels, data filters, and so on. that facilitate your analysis.
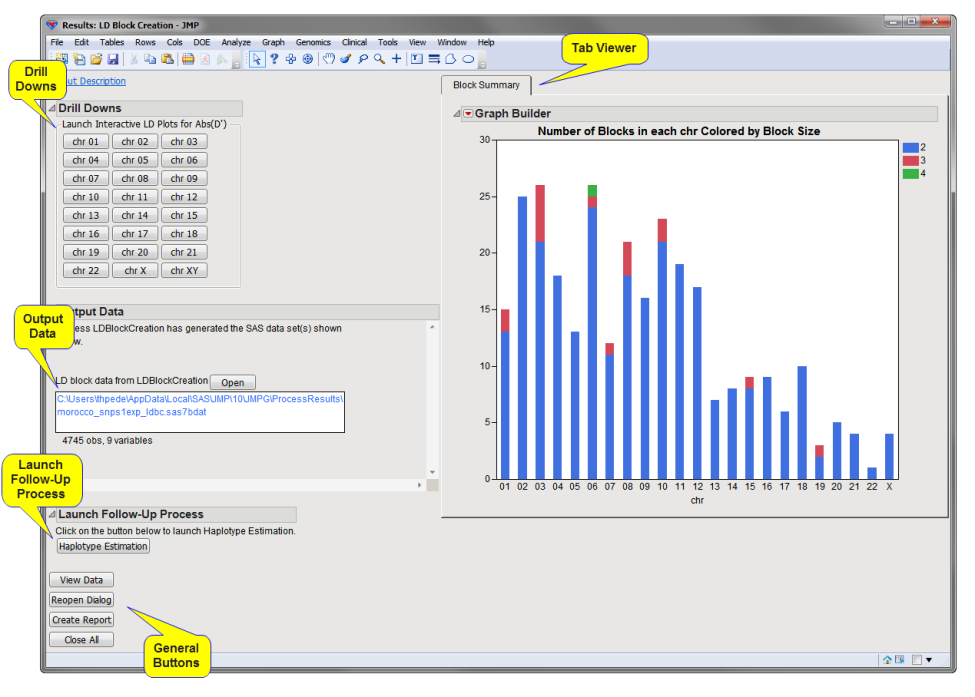
The Results window contains the following panes:
Tab Viewer
This pane provides you with a space to view individual tabs within the Results window. Use the tabs to access and view the output plots and associated data sets.
The following tabs are generated by this process:
| • | Block Summary: This tab shows stacked bar charts of the number of blocks of particular sizes in each chromosome (or other value represented by the annotation group variable), with color representing the size (number of markers in the block). |
Drill Downs
Action buttons provide you with an easy way to drill down into your data. The following action buttons are generated by this process:
| • | : There is one button per annotation group (or one for all markers if no annotation groups defined. Each button opens a window with a HaploView-style LD plot for that annotation group (chromosome, for example) along with a legend displaying the color gradient for the values for Abs(D'). |
The HaploView-style LD plot for chromosome 6 (generated by clicking chr 06,above) is shown below:
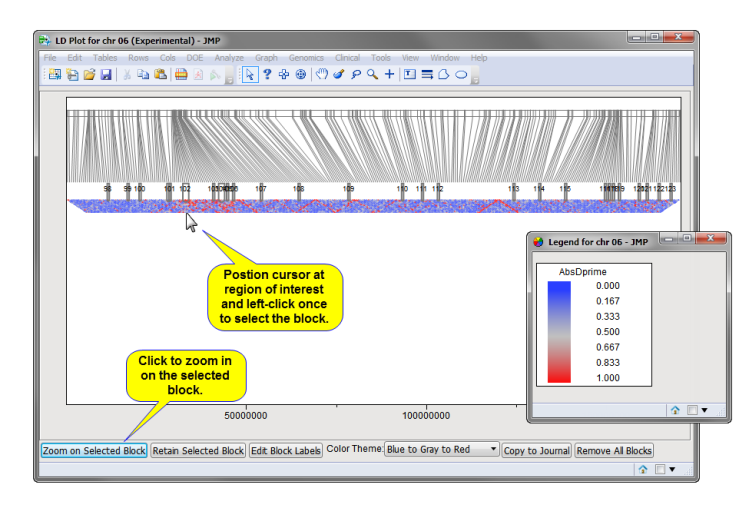
Markers are spaced uniformly in the triangular plot, but have lines drawn to their chromosomal position. LD blocks created by this process are displayed, numbered sequentially across chromosomes by location. You can zoom into selected blocks of interest by selecting the desired block(s) and clicking Zoom on Selected Block (as shown above). The color theme can be changed as well.
The selected region, with several blocks, is shown below.

Output Data
This process generates the following output data set:
| • | Marker Statistics Data Set: This data set contains the columns of the annotation data set along with a Block column indicating what, if any, block a SNP has been assigned to. Click to view the data set. |
For detailed information about the files and data sets used or created by JMP Genomics software, see Files and Data Sets.
General
| • | Click to reveal the underlying data table associated with the current tab. |
| • | Click to reopen the completed process dialog used to generate this output. |
| • | Click to generate a pdf- or rtf-formatted report containing the plots and charts of selected tabs. |
| • | Click to close all graphics windows and underlying data sets associated with the output. |