Output Description
VCF Input Engine
Running this process for the TreoExample sample setting generates the Results window shown below. Refer to the VCF Input Engine process description for more information.
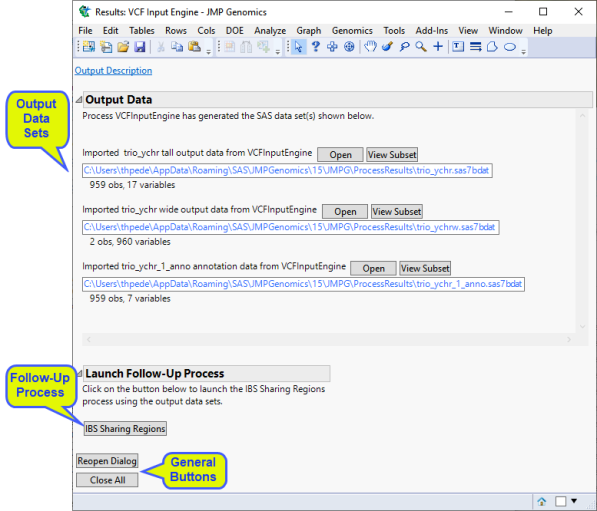
The Results window contains the following elements:
Output Data
This process generates the following data sets:
| • | Output Genotype Data Set (Tall): This data set lists genotype data imported from the VCF file(s) in tall data set format. Refer to Tall and Wide Data Sets for more information about tall data sets. |
| • | Output Genotype Data Set (Wide): This data set lists genotype date imported from the VCF file(s) in wide data set format. Refer to Tall and Wide Data Sets for more information about tall data sets. Note: Wide data sets are generated only when the Create wide Output Genotype Data Sets check box is checked. |
| • | Output Annotation Data Set: This data set lists on the information biological or chemical properties, such as gene identity or chromosomal location, for each of the markers/ molecular entities in the experiment. Refer to Annotation Data Sets for more information about this data set. |
Note: You can view the data in a file or data set by clicking either or (when the data set is very large).
Note: If the genotypes in the GT field of the original VCF files are formatted as "0/0", "1/0", “0/1”, or "1/1". they are transformed into the numeric values 0, 1 or 2, as follows: "0/0", is recoded as “0”, "0/1" and “1/0” are recoded as “1”, and “1/1” is recoded as “2”.
Launch Follow-up Processes
| • | IBS Sharing Regions: Click to launch the IBS Sharing Regions process with the Output Tall Genotype data set specified as input. |
General
| • | Click to reopen the completed process dialog used to generate this output. |
| • | Click to close all graphics windows and underlying data sets associated with the output. |