Process Description
Copy Number/LOH Control Set Adjustment
The Copy Number/LOH Control Set Adjustment process performs copy number intensity or count adjustment by subtraction of control sample intensities or counts, from experimental samples. The subtraction can be done either within subject (for example, tumor-normal paired samples) or across a set or sets of experimental subjects and control subjects.
What do I need?
Two data sets are required for this process:
|
•
|
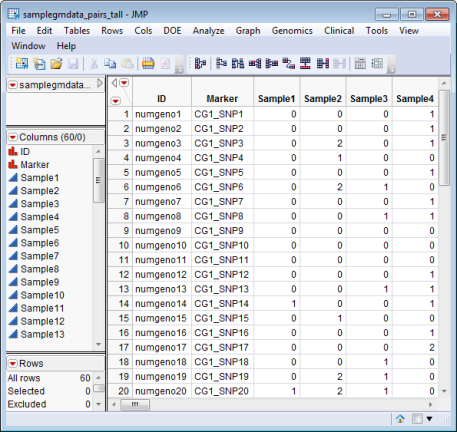
The Input Data Set, containing all of the numeric data to be analyzed. The data set must be in the tall format, with samples or subjects as columns, and copy number measurements as rows. The samplegmdata_pairs_tall.sas7bat data set serves as an example, and is partially shown below.
|
|
•
|
The Experimental Design Data Set (EDDS). This required data set tells how the experiment was performed, providing information about the columns in the input data set. Note that one column in the EDDS must be named ColumnName. The purpose of this column is to index the names of the columns in the input data set; values contained in this column must exactly match the column names in the input data set. An experimental design file with variables that define the experimental conditions of the samples is necessary to set up the sample subtraction. The samplegmdata_pairs_exp.sas7bdat EDDS serves as an example, and is shown below. |

An Annotation Data Set can also be specified. This data set contains information such as gene identity or chromosomal location, for each of the rows in the input data set. This data set is also in the tall format; where each row corresponds to a different molecular entity. The samplemap_numgeno.sas7bdat data set serves as an example, and is shown below.

For this example,
|
•
|
Marker is to be chosen for Variables By Which to Merge Annotation Data. |
|
•
|
The Sample Set(s) to Use for Subtraction are Control Samples Paired with Experimental Samples. |
|
•
|
Ped_id and disease are the Variables Defining Analysis Groups. |
|
•
|
disease is the Variable Defining Control Sets. |
|
•
|
The Control Level is “0”. |
|
•
|
The Control Set Summary Statistic is the mean. |
|
•
|
The Absolute Value of the Differences are to be calculated. |
|
•
|
Only experimental samples are to be output. |
|
•
|
LOH targeted subtraction is to be performed. |
|
•
|
The Control Reference Value is 1. |
|
•
|
Marker is the Annotation Merge Variable. |
|
•
|
Marker, Location, and Candgene are the Annotation Variables to Retain in the Output Data Set. |
For detailed information about the files and data sets used or created by JMP Genomics software, see Files and Data Sets.
Output/Results
Refer to the Copy Number/LOH Control Set Adjustment output documentation for detailed descriptions of the output of this process.


