Process Description
Partition
The Copy Number Partition process uses recursive partitioning to find potential breakpoints of copy number changes across a chromosome. A separate set of breakpoints is computed for each input variable and then the collection of breakpoints is compared graphically.
What do I need?
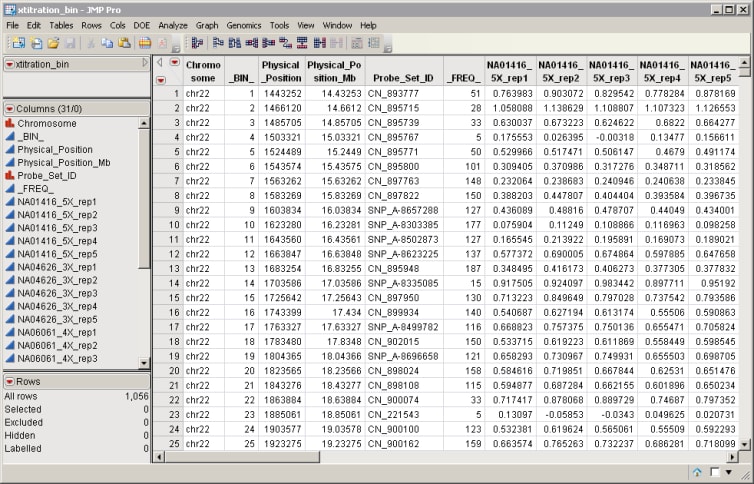
One Input Data Set containing chromosome and position information along with the intensity data is all that is required to run this process. An example is the xtitration_bin.sas7bdat.sas7bdat data set shown below.
Caution and Tip: Processing hundreds or thousands of variables can result in long run times. To save processing time, you should consider binning vary large data sets, using the Bin process, before running this process. Alternatively, you might wish to run the One-Way ANOVA process first to identify chromosomes that display interesting copy number differences between experimental groups, or when individuals are compared to a control group. You can then specify the chromosomes of interest in the Filter to Include Observations field on the General tab to restrict your partition analysis to only those chromosomes.
This data set has been binned using the Bin process. Note:
| • | Bin identifiers are listed in column 2. |
| • | Columns 1 and 3 list chromosome and positional information, respectively. |
| • | Intensity data are listed in columns 8 through 31. |
An Experimental Design Data Set (EDDS) is optional. This data set tells how the experiment was performed, providing information about the columns in the input data set. Note that one column in the EDDS must be named ColumnName. The purpose of this column is to index the names of the columns in the input data set; values contained in this column must exactly match the column names in the input data set. The xtitration_edf.sas7bdat EDDS serves as an example.

In this example,
| • | NA01416_5X_rep1 through NA15510_2X_fosmid_rep5 have been selected as Variables to Partition Intensity Values, |
| • | Chromosome has been selected as the Chromosome Variable, and |
| • | Physical_Position has been selected as the Position Variable. |
For detailed information about the files and data sets used or created by JMP Genomics software, see Files and Data Sets.
Output/Results
The output generated by this process is summarized in a Tabbed report. Refer to the Partition output documentation for detailed descriptions and guides to interpreting your results.