Process Description
Malecot LD Map
A useful method for describing the variations in patterns of LD across large DNA regions involves calculating the metric linkage disequilibrium units (LDUs) between pairs of SNPs (Maniatis et al., 2002). The LDU scale, which has additive distances and locations monotonic with physical and genetic maps (Zang, W. et al. 2002), provides a coordinate system whose scale is proportional to the regional differences in the strength of LD, in a fashion analogous to the recombination maps constructed in cM used to guide linkage studies (Kong, A., et al., 2002). A pattern of plateaus, which correspond to regions of high LD, and steps, which correspond to regions of increased recombination (Zang, W. et al. 2002), is evident.
The Malecot LD Map process uses an estimate of the LD measure Rho (calculated using the Linkage Disequilibrium process and included in the output data set ending with _ldstats that it creates) to fit the Malecot model Rho = (1-L)*M*exp(-E_i*d_i)+L to each interval of length d_i kb between consecutive SNPs via nonlinear optimization routines. Distances between pairs of SNPs can then be represented in terms of LD units (LDU), which are computed by summing (E_i*d_i) over all intervals between the pair of SNPs. The distance in terms of LDU is plotted versus the physical distance between consecutive SNP pairs in the plot(s). For reference, see Maniatis et al. (2002).This Analytical Procedure uses SAS PROC NLMIXED to fit the Malecot model by weighted nonlinear least squares.
What do I need?
Before running the Malecot LD Map process, an intermediate data set must be created using either the Linkage Disequilibrium process or another program that creates a data set containing variables with the following information for each pair of genetic markers (for example, SNPs):
| 1 | a label for each pair of markers, |
| 2 | the physical location of each marker, and |
| 3 | the values of rho and K_rho for the pair. |
The samplegmdata data set used in the following example was computer generated and consists of 1000 rows of individuals with 130 columns corresponding to data on these individuals. This is a wide data set; markers are listed in columns, whereas individuals are listed in rows. The samplegmdata data set is included in the Sample Data folder that comes with JMP Genomics and is described in Data Sets Used in JMP Genomics Processes.
The intermediate samplegmdata.ldstats data set was generated using the Linkage Disequilibrium process. The data set has 16 columns and 930 rows. The Estimate Rho and K_Rho box found on the Options tab of the Linkage Disequilibrium dialog was checked. This samplegmdata.ldstats data set, illustrated below, was saved in the ProcessResults folder.
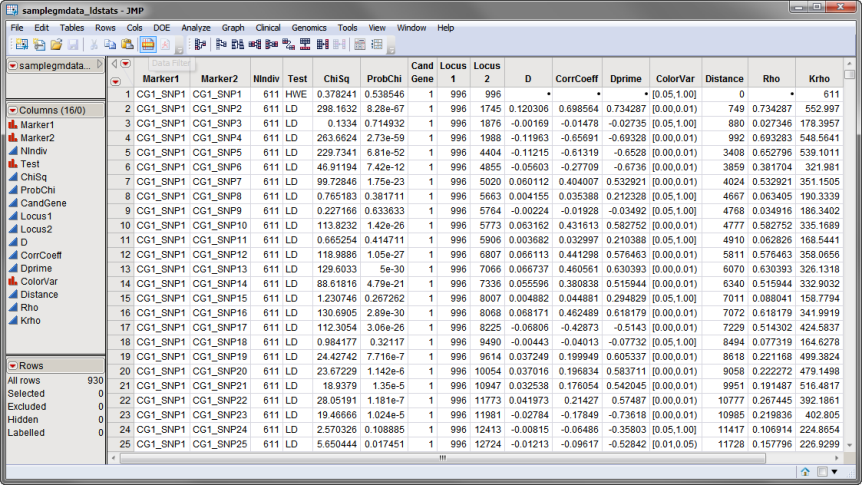
The samplegmdata_ldstats.sas7bdat data set contains the elements needed for successful use of the Malecot LD Map process. The labels for each pair of SNPs are contained in the Marker1 and Marker2 columns. The location for each of the SNPs is given in the Locus1 and Locus2 columns. Finally, the values for Rho and K_Rho are given in the Rho and Krho columns.
For detailed information about the files and data sets used or created by JMP Genomics software, see Files and Data Sets.
Output/Results
The output generated by this process is summarized in a Tabbed report. Refer to the Malecot LD Map output documentation for detailed descriptions and guides to interpreting your results.