Process Description
Affymetrix Cytogenetics/CytoscanHD CEL Input Engine
The Affymetrix Cytogenetics/CytoscanHD CEL Input Engine imports a set of .cel files generated from Cytogenetics 2.7M .cel or CytoScan HD chip files and combines them into a single SAS data set.
What do I need?
Before you can successfully import the raw data into SAS data sets that can be used for analysis in JMP Genomics, you must locate and gather several sources of information:
| • | An Experimental Design File (EDF) that indexes the individual .cel files for the experiment. The EDF is typically a text file or Excel spread sheet and must be created before the data can be imported. Alternatively, you can generate an EDF by parsing the .arr files, associated with each .cel file, using the Affymetrix ARR File Parser process. The ARR File Parser is a JMP Scripting Language(JSL) script that parses information from all of the .arr files into two JMP files. The first output file (named ARR.jmp, by default) contains all the experimental parameters listed in all of the .arr files. The second output file (named EDF.jmp, by default) lists only those variables that vary across chips. This EDF.jmp file can be modified to serve as an EDF for subsequent JMP Genomics processes. Please refer to Affymetrix ARR File Parser for more information about using the ARR File Parser process. |
| • | All of the .cel files containing the raw data. They must be located and copied to a single folder. Each .cel file corresponds to an individual microarray and contains the hybridization intensities for that array. |
| • | A SNP Annotation data set. (Another optional annotation data set, the Copy Number Annotation data set, can also be included.) These data sets contain annotation information imported from the Affymetrix .CSV annotation file for the specific SNP GeneChips used in the experiments. Each row in the SNP Annotation data set corresponds to a separate SNP probeset. (Each row in the Copy Number Annotation data set corresponds to a separate copy number probeset.) Required variables for both data sets include Probe_Set_ID, Chromosome, and Physical_Position. To generate each of these data sets, first download the specific .CSV file from the Affymetrix website, and then import the data into a SAS data set using the Import Individual Text, CSV, or Excel Files process. |
The following example uses a subset of the Chromosome Analysis Suite (ChAS) sample data available from Affymetrix. The data includes five sets of cytogenetics sample data generated on the Cytogenetics Whole Genome 2.7 Array. Each set includes three files: a .cel file, an .arr file and a .CYCHP file. Files were downloaded from Affymetrix and saved in an AffyCytogenetics folder created in the JMP Genomics Sample Data folder.

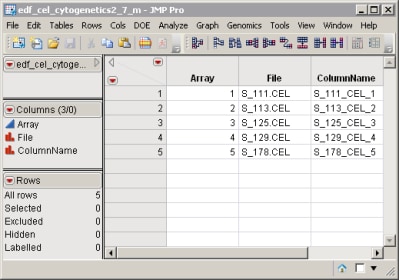
The EDF was generated using information contained in the .arr files associated with each of the raw data files. Files were parsed using the Affymetrix ARR File Parser process. The required ColumnName variable was added to the resulting EDF.jmp file using the Create ColumnName process and the edf_cel_cytogenetics2_7_m modified EDF was saved as a SAS data set in the AffyCytogenetics folder.
The Cytogenetics_Array.cdf file was downloaded from the Affymetrix website and saved in the AffyCytogenetics folder.
The _cytogenetics_array_na29_annot.sas7bdat SNP annotation file was generated by importing .csv files, downloaded from NetAffx, using the Import Individual Text, CSV, or Excel Files process. The resulting SAS data set was saved in the AffyCytogenetics folder.

No copy number annotation data set is used in this example.
For detailed information about the files and data sets used or created by JMP Genomics software, see Files and Data Sets.
Output/Results
The output data sets generated by this process are listed in a Results window. Refer to the Affymetrix Cytogenetics/CytoscanHD CEL Input Engine output documentation for detailed descriptions.