Process Description
Affymetrix CNAT Input Engine
The Affymetrix CNAT Input Engine imports multiple CNAT4.0 files into a single SAS data set, with rows representing individual SNPs, and columns containing various copy number or loss-of-heterozygosity measurements, for each of the samples.
What do I need?
Before you can successfully import the raw data into SAS data sets that can be used for analysis in JMP Genomics, you must locate and place all of the .CNT files, output by the CNAT process, into a single folder.
Each .CNT file is a tab-delimited text file and corresponds to an individual microarray. Its header contains specific information about the format of the chip. Below the header are columns identifying specific spots along with their copy number (CN) or loss-of-heterozygosity (LOH) estimates and statistics. An example of one such file is shown below.
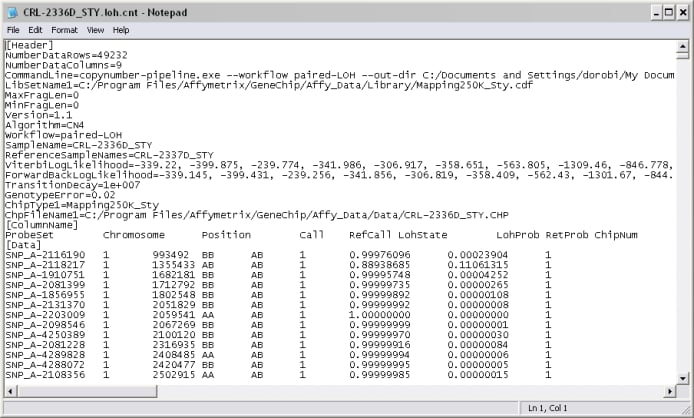
.CNT files typically occur in pairs. Both files in a pair share the same name but have different suffixes appended. One file in the pair lists copy number (CN) data and has the .cn.cnt suffix. The other lists loss-of-heterozygosity (LOH) data and has the .loh.cnt suffix.
The following example uses a subset of the 500K HapMap Normal data set available for download from Affymetrix. Compressed files containing LOH and CN data from three samples, each on both Nsp and Sty arrays, were downloaded from the Affymetrix website, unzipped, and saved to a folder named CNAT in the Genomics\Sample Data folder. Included files are listed below.

For detailed information about the files and data sets used or created by JMP Genomics software, see Files and Data Sets.
Output/Results
The output data sets generated by this process are listed in a Results window. Refer to the Affymetrix CNAT Input Engine output documentation for detailed descriptions.