Process Description
Combine Columns
The Combine Columns process groups together columns of a tall data set and computes a summary statistic, such as the mean for each group. It also creates an accompanying reduced Experimental Design Data Set (EDDS). This process is useful when you want to combine similar columns of data (for example, from replicate runs) together into one column.
What do I need?
Two SAS data sets are required for the Combine Columns process:
| • | The Input Data Set containing the columns to be reordered. The drosophilaaging_norm.sas7bdat data set (located in the \LifeSciences\Sample Data\Microarray\Scanalyze Drosophila directory included with JMP Genomics, associated with the Drosophila aging experiment of Jin et al. (2001) described in Drosophila Aging Experimental Data) serves as an example. |
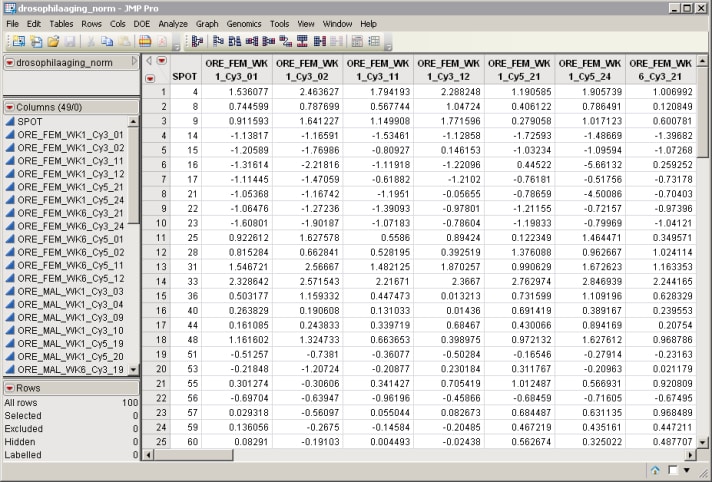
| • | An Experimental Design Data Set (EDDS) that tells how the experiment was performed, providing information about the columns of the primary experimental data. The drosophilaaging_exp.sas7bdat EDDS (located in the \LifeSciences\Sample Data\Microarray\Scanalyze Drosophila directory included with JMP Genomics, associated with the Drosophila aging experiment of Jin et al. (2001) described in Drosophila Aging Experimental Data) serves as an example. |
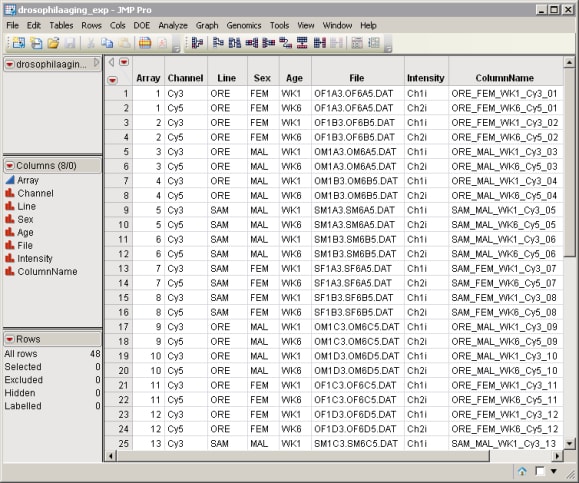
To follow this example, complete the following steps in the dialog:
| 8 | Specify Line, Sex, and Age as Variables Defining Groups. |
| 8 | Select Mean from the Statistic to Compute drop-down menu. |
For detailed information about the files and data sets used or created by JMP Genomics software, see Files and Data Sets.
Output/Results
The output of the Combine Columns process includes the Results window, which lists the output data sets:
| • | The output data set, and |
| • | The output EDDS. |
| 8 | Click to view the drosophilaaging_norm_gc.sas7bdat output data set. |
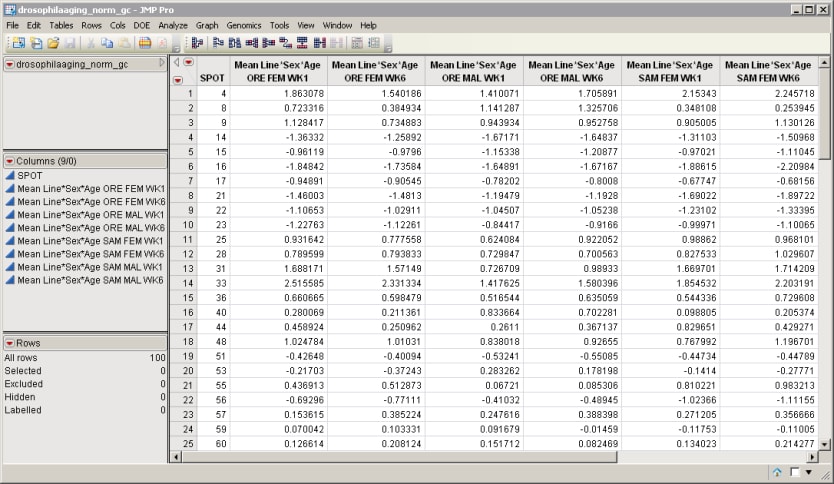
The output data set contains nine columns (one for each combination of line, sex, and age) and 100 rows. Each group of six columns, matching the specified criteria from the input data set, were combined into one column. No rows were eliminated. Instead of raw intensities, each column lists the mean for each group of combined rows.
| 8 | Click to view the drosophilaaging_exp_gc.sas7bdat output EDDS. |

The output EDDS contains the same nine columns as the input EDDS but only eight rows. Each row corresponds to one column from the drosophilaaging_norm_gc.sas7bdat output data set.
These files are now ready for further analysis.