Bivariate Fit Options
The Bivariate platform model-specific red triangle options depend on the specified fit. These menus are found next to the model fit label under the scatterplot.
• Options That Apply to Most Bivariate Fits
• Options That Apply to Normal Ellipses
• Options That Apply to Quantile Density Contours
Options That Apply to Most Bivariate Fits
The Bivariate platform model-specific red triangle options depend on the specified fit. Not all options are available for all fits.
Line of Fit
Shows or hides the line, curve, or contours that describe the model fit. Not applicable for Quantile Density Contours.
Confid Curves Fit
Shows or hides the confidence limits (curves) for the expected value (mean).
Confid Curves Indiv
Shows or hides the confidence limits for an individual predicted value. The confidence limits reflect variation in the error and variation in the parameter estimates.
Line Color
Enables you to select a color for the fit curve.
Line Style
Enables you select a line style for each fit.
Line Width
Enables you to select a line width for each fit.
Report
Shows or hides the report for each fit.
Note: This option does not modify the Bivariate plot.
Profiler
Shows or hides a prediction trace for the X variable. Enables you to find optimum settings for one or more responses and to explore response distributions using simulation. See Profiler in Profilers.
Save Predicteds
Creates a new column in the current data table called Predicted <colname> where colname is the name of the Y variable. This column contains the prediction formula and the predicted values.
Tip: The prediction formula computes values automatically for new rows that you add to the table.
Save Residuals
Creates a new column in the current data table called Residuals <colname> where colname is the name of the Y variable. This column contains the observed response values minus their predicted values.
Note: You can use the Save Predicteds and Save Residuals options for each fit. If you use these options multiple times or with a grouping variable, it is best to rename the resulting columns in the data table to reflect each fit.
Save Studentized Residuals
Creates a new column in the current data table called Studentized Residuals <colname> where colname is the name of the Y variable. This column contains the residuals divided by their standard errors.
Mean Confidence Limit Formula
Creates two new columns in the data table called Lower 95% Mean <colname> and Upper 95% Mean <colname> where colname is the name of the Y variable. These columns contain both the formulas and the values for lower and upper 95% confidence limits for the mean response.
Indiv Confidence Limit Formula
Creates two new columns in the data table called Lower 95% Indiv <colname> and Upper 95% Indiv <colname> where colname is the name of the Y variable. These columns contain both the formulas and the values for lower and upper 95% confidence limits for an individual prediction.
Plot Residuals
(Available for Linear, Polynomial, Passing Bablok, and Fit Special Only.) Shows or hides five diagnostic plots: residual by predicted, actual by predicted, residual by row, residual by X, and a normal quantile plot of the residuals.
Bland Altman Analysis
(Available only for Passing Bablok.) Shows a matched pairs and Bland-Altman analysis in a new report window.
Set α Level
Enables you to specify the alpha level used in computing confidence curves.
Confid Shaded Fit
(Not available for all fits.) Shows or hides a shaded confidence region for the expected response (mean).
Confid Shaded Indiv
(Not available for all fits.) Shows or hides a shaded confidence region for an individual prediction.
Save Coefficients
(Available only for flexible spline fits.) Saves the spline coefficients as a new data table that contains columns named X, A, B, C, and D. The X column contains the knot points. A, B, C, and D are the intercept, linear, quadratic, and cubic coefficients of the third-degree polynomial. These coefficients span from the corresponding value in the X column to the next highest value.
Remove Fit
Removes the fit from the graph and removes its report.
Options That Apply to Normal Ellipses
The Normal Ellipse red triangle options are available for Density Ellipse fits in the Bivariate platform.
Shaded Contour
Shades or unshades the area inside the density ellipse.
Select Points Inside
Selects the points inside the ellipse.
Select Points Outside
Selects the points outside the ellipse.
Options That Apply to Quantile Density Contours
The Quantile Density Contours red triangle options are available for Nonpar Ellipse fits in the Bivariate platform.
Kernel Control
Shows or hides a slider to control the standard deviation for each variable. The standard deviation defines the range of X and Y values for determining the density of contour lines.
5% Contours
Shows or hides the 5% contours.
Contour Lines
Shows or hides the contours.
Contour Fill
Shows or hides filled contours.
Color Theme
Enables you to change the color theme of the contours.
Select Points by Density
Enables you to select points that fall in a user-specified quantile range.
Color by Density Quantile
Colors the points according to density.
Save Density Quantile
Creates a new column in the current data table that contains the density quantile for each point.
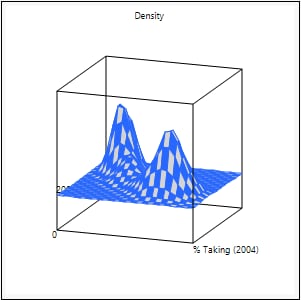
Mesh Plot
Shows or hides a three-dimensional plot of the density over a grid of the two analysis variables.
Figure 5.6 Example of a Mesh Plot
Modal Clustering
Shows or hides the results for a modal clustering of the data. Creates a new column in the current data table that contains the cluster number for each data pair. Modal clustering is based on the density estimates. JMP generates a grid of 10,404 density estimates. The number of modes in the distribution of the densities determines the number of clusters. The modes are the cluster centers. The remaining points are iteratively clustered to each mode based on distance and density estimates.
Note: For more information about additional clustering methods in JMP, see Overview of Platforms for Clustering Observations inMultivariate Methods.
Save Density Grid
Creates a new data table that contains the density estimates and the quantiles associated with them.
Note: If you save the modal clustering values first and then save the density grid, the grid table also contains the cluster values.