Additional Example of Bootstrapping
This example illustrates the benefits of the Fractional Weights (Bayesian Bootstrap) option for a small data table. The data consist of a response, Y, measured on three samples of each of seven different soil types. A scientist is interested in finding a confidence interval for the mean response for the wabash soil type.
Because each soil type has only three observations, the simple bootstrap has the potential to exclude all three of the observations for wabash from a bootstrap sample. The Fractional Weights option ensures that all observations for every soil type are represented in all bootstrap samples.
The scientist examines the distribution of wabash sample means from both bootstrap methods:
Simple Bootstrap Analysis
1. Select Help > Sample Data Folder and open Snapdragon.jmp.
2. Select Analyze > Fit Y by X.
3. Select Y and click Y, Response.
4. Select Soil and click X, Factor.
5. Click OK.
6. Click the red triangle next to Oneway Analysis of Y By Soil and select Means/Anova.
7. In the Means for Oneway Anova report, right-click the Mean column and select Bootstrap.
8. Type 1000 for the Number of Bootstrap Samples.
9. (Optional) To match the results in Figure 11.7, type 12345 for the Random Seed.
10. Click OK.
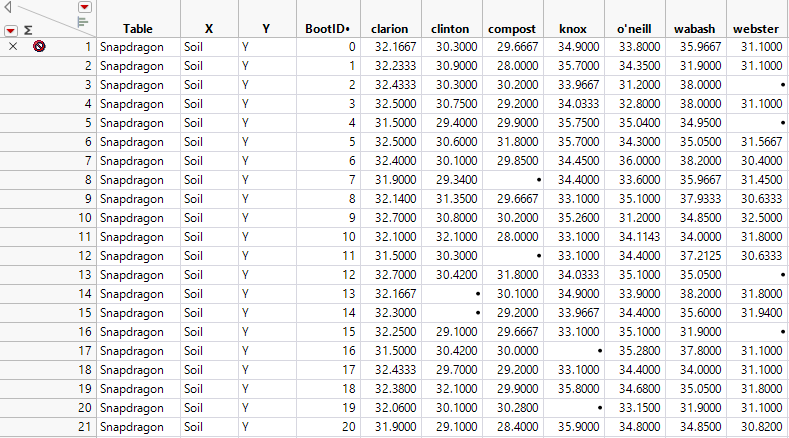
Figure 11.7 Bootstrap Results for a Simple Bootstrap
The missing values in Figure 11.7 represent bootstrap iterations in which none of the observations for a given soil type were selected for the bootstrap sample.
11. Select Analyze > Distribution.
12. Select wabash and click Y, Columns.
13. Click OK.
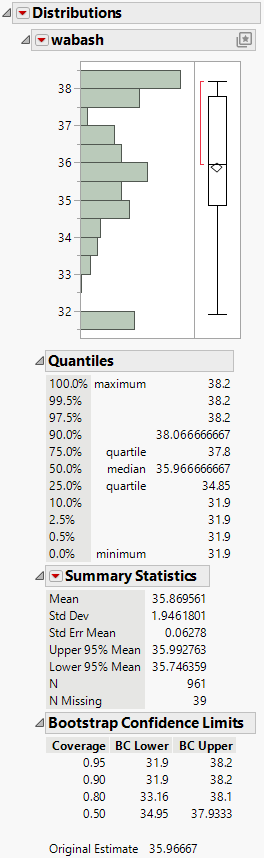
Figure 11.8 Distribution of wabash Means from a Simple Bootstrap
Figure 11.8 shows the distribution of wabash means from the simple bootstrap analysis. Notice the following:
– The Summary Statistics report indicates that the number of rows containing bootstrap means for wabash is N = 961. Although you conducted 1,000 iterations, 39 bootstrap samples did not contain any of the three observations for wabash.
– The histogram of sample means is not smooth, with peaks at the two extremes. The three values for wabash are 38.2, 37.8, and 31.9. The peak at the low end of the distribution results from bootstrap samples that contain only the value 31.9. The peak at the high end results from bootstrap samples that contain one or both of the values 38.2 and 37.8.
Next, use the Fractional Weights (Bayesian Bootstrap) option to avoid obtaining missing values from the bootstrap samples and to smooth the distribution of bootstrapped means.
Bayesian Bootstrap Analysis
1. In the Oneway Analysis report, right-click the Mean column in the Means for Oneway Anova report and select Bootstrap.
2. Type 1000 for the Number of Bootstrap Samples.
3. (Optional) To match the results in Figure 11.9, type 12345 for the Random Seed.
4. Select the Fractional Weights option.
5. Click OK.
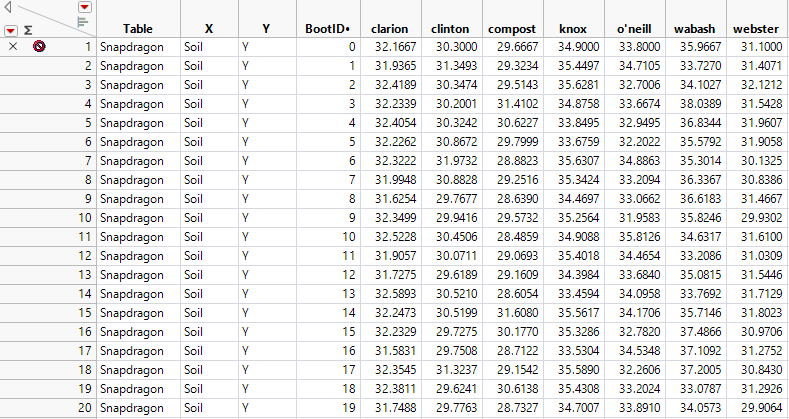
Figure 11.9 Bootstrap Results for a Bayesian Bootstrap
There are no missing values in the Bayesian Bootstrap results table. All 21 rows in the Snapdragon.jmp data table are included, with varying bootstrap weights, in each bootstrap sample.
6. Select Analyze > Distribution.
7. Select wabash and click Y, Columns.
8. Click OK.
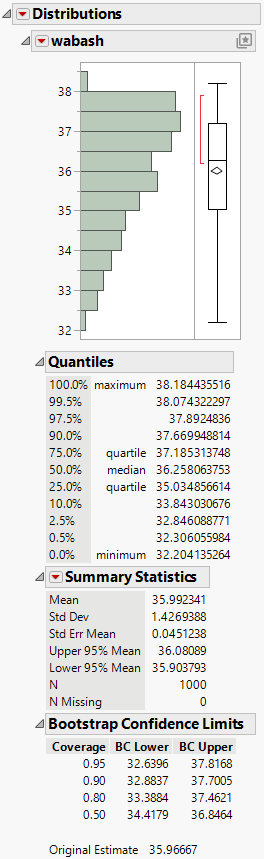
Figure 11.10 Distribution of wabash Means from a Bayesian Bootstrap
The Bayesian Bootstrap produces a much smoother distribution for the wabash sample means. All 1,000 bootstrap samples include the three observations for wabash. For each iteration, the wabash sample mean is calculated using different fractional weights.
The Bootstrap Confidence Limits report shows that a 95% confidence interval for the mean is 32.6396 to 37.8168.