Example of Augmenting a Nonlinear Design
Create a nonlinear design when you have prior data. In this example, you want to model the rate of uptake (velocity) of available organic substrate as a function of the concentration of that substrate. You have already run an experiment, but you want to leverage your results to obtain more precise estimates of the parameters.
Obtain Prior Parameter Estimates
Use your existing experimental data to obtain better estimates of the parameter values.
1. Select Help > Sample Data Folder and open Nonlinear Examples/Chemical Kinetics.jmp.
2. Click the plus sign next to Model (x) in the Columns panel. The formula editor opens.
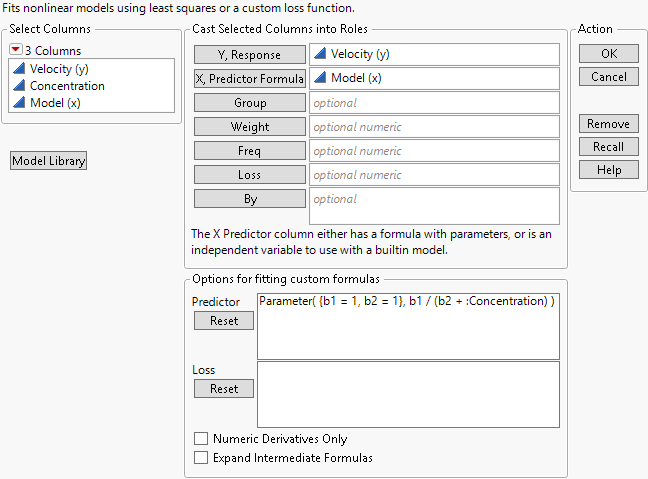
3. TheParameters section in the middle bottom of the formula editor shows the current values of the model parameters.The values (b1 = 1 and b2 = 1) are your initial estimates. They are used to compute the Model (x) values in the data table. For your next experiment, you want to replace these with better estimates.
4. Click Cancel to close the formula editor window.
5. Select Analyze > Specialized Modeling > Nonlinear.
6. Select Velocity (y) and click Y, Response.
7. Select Model (x) and click X, Predictor Formula.
Notice that the formula given by Model (x) appears in the Options for fitting custom formulas panel.
Figure 23.8 Nonlinear Analysis Launch Window
8. Click OK.
9. In the Control Panel, click Go.
The iterative search for a solution proceeds until one of the Stop Limit values is reached. Then, the Solution and Correlation of Estimates reports appear. Also, an option appears in the Control Panel enabling you to add confidence limits to the Solution report.
10. In the Control Panel, click Confidence Limits.
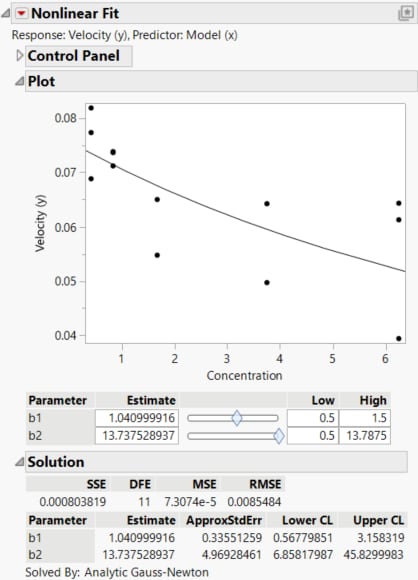
Confidence intervals for the parameters b1 and b2 appear in the Solution report.
Figure 23.9 Nonlinear Fit Results
The Lower CL and Upper CL values for b1 and b2 define ranges of values for these parameters. Next, use these intervals to define a range for the prior values in your augmented nonlinear design.
Note: Do not close the Nonlinear Fit report because these results are needed in the next steps.
Augment the Design
Now, create a design to estimate the nonlinear parameters more precisely.
1. With the Chemical Kinetics.jmp data table active, select DOE > Special Purpose > Nonlinear Design.
2. Select Velocity (y) and click Y, Response.
3. Select Model (x) and click X, Predictor Formula.
4. Click OK.
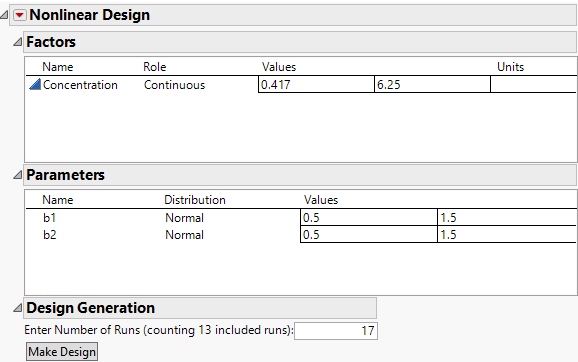
Figure 23.10 Nonlinear Design Sections for Factors and Parameters
In the Chemical Kinetics.jmp data, the values for Concentration range from 0.417 to 6.25. Therefore, these values initially appear as the low and high values in the Factors section. You want to change these values to encompass a broader interval.
5. Click 0.417 and type 0.1. Click 6.25 and type 7.
6. Leave the prior Distribution for each parameter set to Normal.
The range of Values for the parameters reflects the uncertainty of your knowledge about them. You could specify a range that you think covers 95% of possible parameter values. The confidence limits from the Nonlinear Fit report shown in Figure 23.9 provide such a range. Replace the Values for the parameters in the Parameters section with the confidence limits, rounding to three decimal places.
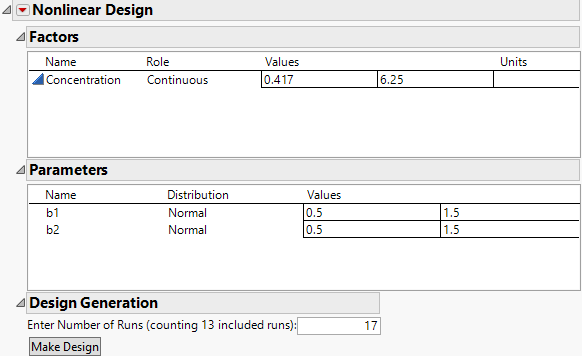
7. In the DOE Nonlinear Design window, enter these values into the Parameters for b1 and b2:
– b1: 0.568 and 3.158
– bb2: 6.858 and 45.830
Figure 23.11 Updated Values for Factor and Parameters
8. Enter 40 for the Number of Runs in the Design Generation panel.
9. Click Make Design.
The Design section opens, showing the Concentration and Velocity (y) values for the original 13 runs and new Concentration settings for the additional 27 runs.
10. Click Make Table.
This creates a new JMP design table that contains the settings and results for the original 13-run design and settings for 27 new runs. Instead of creating a new data table, you can add the new runs to your existing data table by clicking Augment Table instead of Make Table.
The new runs reflect the broader interval of Concentration values and the range of values for b1 and b2 obtained from the original experiment, which are used to define the prior distribution. Both should lead to more precise estimates of b1 and b2.