Example of Marker Relatedness
Learn how to simulate genetic crosses from your genetic data.
1. Select Help > Sample Data Folder > Life Sciences and open Genotypes Pedigree.jmp and Genotypes Pedigree Anno.jmp.
2. Click on the Genotypes Pedigree.jmp table.
3. Select Analyze > Genetics > Marker Relatedness.
4. Select Markers (60/0) and click Marker.
5. Select Sex and click By.
6. Enter 2 for Ploidy.
7. Enter 12345 for Set Random Seed.
8. Set Kinship Type to Additive.
9. Specify Diploid Method 1 for the Additive Type.
10. Set Missing Marker Imputation Method to Random.
11. Leave both the Principal Components and Clustering boxes checked.
12. Click OK.
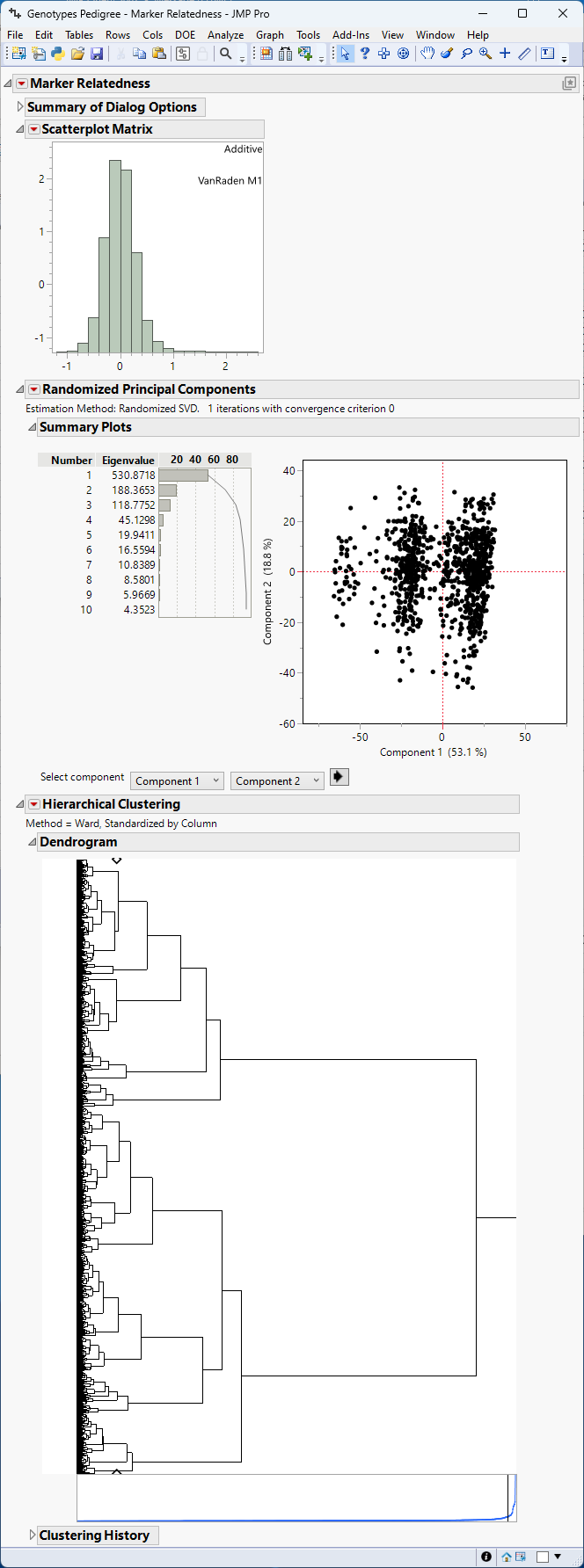
The Marker Relatedness report is generated as shown in Figure 5.1.
Figure 5.2 Relatedness of the markers
The Marker Relatedness report was run once for each sex (M and F) and contains the following information for each sex. The results are shown for one sex only.
• The Summary of Dialog Options summarizes the selections made on the platform and the time it took for the simulation to run.
• The Scatterplot Matrix display shows the histogram (and correlation plots when estimating epistasis). A histogram skewed to the right (high values) indicate that most individuals have strong genetic similarity. On the contrary, a histogram skewed to the left (low values) indicate that most individuals have weak genetic similarity.
• The Principal Components Plot shows the clusters of individuals grouped by their relatedness. In the example shown here, the samples do not appear to form definite groups.
• The Clustering Plot is another way to view groups of related individuals. In the red triangle, click on the option Two Way Clustering to produce a head map of relationship estimates among individuals, which is very useful to visualize the range of estimates graphically among all pairs of individuals.