Launch the Marker Relatedness Platform
To estimate a square matrix of genetic relationship between pair of individuals in the data table, launch the Marker Relatedness platform by selecting Analyze > Genetics > Marker Relatedness.
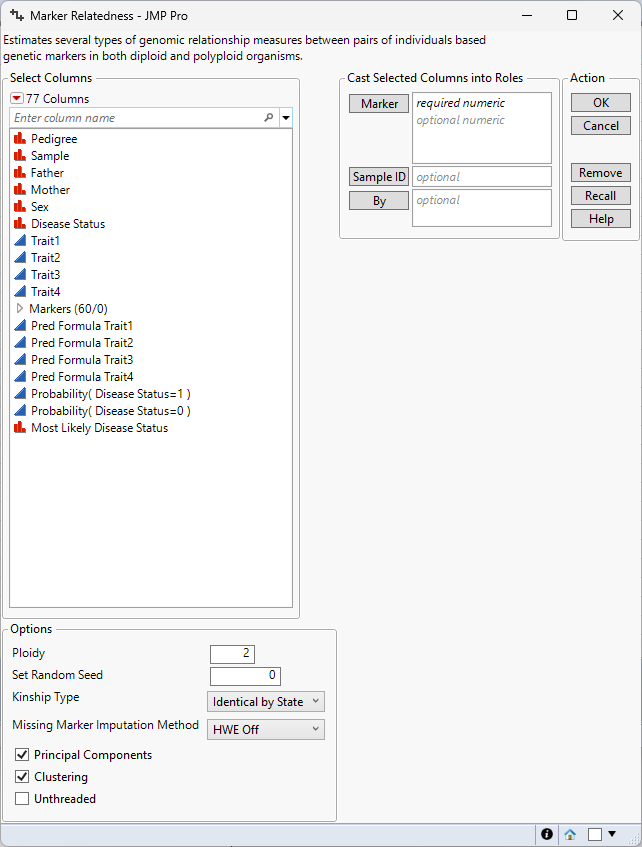
Figure 5.3 Marker Relatedness Launch Window
Marker
Select desired marker columns and click Marker to specify the markers that you want to analyze.
Sample ID
Use this option to specify one variable whose values that can provide a unique identifier for each row.
Note: Only one variable is allowed.
By
Produces a separate report for each level of the By variable. If more than one By variable is assigned, a separate report is produced for each possible combination of the levels of the By variables.
Ploidy
Enables you to specify the ploidy level of the experimental organism under investigation. Note: This must be an even number
Set Random Seed
Use this option to specify a nonnegative integer to start the random number stream. Different values produce different outcomes of the algorithm.
Kinship Type
Use this option to specify the type of measurement to use to assess the relationship between samples (rows).
• Select Identical by State to examine the effects of marker identity on relatedness of the individuals.
• Select Additive to examine the additive marker effects on relatedness of the individuals.
• Select Dominance to examine the marker dominance effects on relatedness of the individuals.
• Select Epistasis to examine the effect of marker interaction on relatedness of the individuals.
Additive Type
Use this option to specify a method to assess
• Select Diploid Method 1 to compute the relationship matrix using Van Rayden’s first method (Van Raden, et al., 2008). (P.M. Van Raden, “Efficient Methods to Compute Genomic Predictions“. J.Dairy Sci. 91:4414-4423, 2008)
• Select Diploid Method 2 to use the second method of Van Raden, et al., 2008. (P.M. VanRaden, “Efficient Methods to Compute Genomic Predictions“. J.Dairy Sci. 91:4414-4423, 2008)
• Select Polyploid to use the method of Batista et al. (2022). This method builds a covariance matrix of additive effects using estimates of ploidy and allele dosage. Batista, L.G., Mello, V.H., Souza, A.P. et al. Genomic prediction with allele dosage information in highly polyploid species. Theor Appl Genet 135, 723–739 (2022). https://doi.org/10.1007/s00122-021-03994-w
Note: This option is available only when Additive is chosen as the kinship type or when Epistasis is selected AND either Additive by Additive or Additive by Dominance is selected as the Epistatis Type.
Dominance Type
Use this option to specify a method to assess
• Select Diploid Method 1 to use the method of Su, et al. (2012). (Su G, Christensen OF, Ostersen T, Henryon M, Lund MS. Estimating additive and non-additive genetic variances and predicting genetic merits using genome-wide dense single nucleotide polymorphism markers. PLoS One. 2012;7(9):e45293. doi: 10.1371/journal.pone.0045293. Epub 2012 Sep 13. PMID: 23028912; PMCID: PMC3441703.)
• Select Diploid Method 2 to use the method of Vitezica, et al. (2012). This method uses a matrix of dominant genomic relationships across individuals. This matrix can be used in a mixed-model context to estimate dominant variances in the population. Zulma G Vitezica, Luis Varona, Andres Legarra. 2013. On the Additive and Dominant Variance and Covariance of Individuals Within the Genomic Selection Scope, Genetics 195: 1223–1230. (https://doi.org/10.1534/genetics.113.155176)
• Select Polyploid to use the method of Batista et al. (2022). This method builds a covariance matrix of dominance effects using estimates of ploidy and allele dosage. Batista, L.G., Mello, V.H., Souza, A.P. et al. Genomic prediction with allele dosage information in highly polyploid species. Theor Appl Genet 135, 723–739 (2022). https://doi.org/10.1007/s00122-021-03994-w
Note: This option is available only when Dominance is chosen as the kinship type or when Epistasis is selected AND either Additive by Dominance or Dominance by Dominance is selected as the Epistatis Type.
Epistasis Type
Use this method to specify a method to assess
• Select Additive by Additive to compute the interaction between the additive type specified in the dialog with itself. For example,, when Additive Type is Method 1, it computes the interaction between this relationship type with itself.
• Select Additive by Dominance to compute the interaction between the additive and dominance types specified in the dialog. For example,, when Additive Type is Method 1 and Dominance Type is Method 1, it computes the interaction between these two relationship types.
• Select Dominance by Dominance to compute the interaction between the dominance type specified in the dialog with itself. For example,, when Dominance Type is Method 1, it computes the interaction between this relationship type with itself.
Note: This option is available only when Epistasis is chosen as the kinship type
Missing Marker Imputation Method
This platform does not run when your data is missing marker data. Because of this, any missing data must be imputed. Use this option to specify how the missing values are to be imputed.
– Select HWE Off to impute the missing genotypes with random draws from a multinomial distribution in which the frequency of each genotype class is set to be the observed frequency from the data.
– Select HWE On to impute the missing genotypes with random draws from a multinomial distribution in which the frequency of each genotype class is set to be the expected frequency under the assumption of Hardy-Weinberg Equilibrium.
– Select Random to randomly assign one of the acceptable values (0, 1, 2, ..., K (where K is the ploidy level)).
– Select Specified to impute the missing genotypes with a specified integer between zero and the ploidy number.
Imputation Value
Specify a value between 0 and the ploidy number in the text box when you select the Specified option in Missing Value Imputation Method.
The values listed here assume diploid organisms; adjust values accordingly for organisms with higher ploidy.
Principal Components
Check this box to compute and plot principal components on the square matrix.
Clustering
Check this box to cluster the individuals by marker relatedness.
Unthreaded
Suppresses multi-threading. Deselect this option for improved computational speed.
Required Data Format for the Marker Relatedness Platform
Most of the processes in JMP assume that the input table has a particular data structure. JMP distinguishes between tall and wide data sets. A tall data table has samples as columns and molecular entity (for example, marker, gene, clone, protein, or metabolite) as rows, whereas a wide data table is the transpose of the tall data table, having the samples as rows and molecular entity as columns.
When specifying the input data set for a process, it is important to know the required form. Marker Relatedness requires a wide data table. The Transpose platform under the Tables menu enables you to transform your data tables between tall and wide forms.
Marker data must be encoded in the one-column, numeric format. Typically, in this format, diploid individuals homozygous for the least common, or minor allele, are represented in the table by a 2, whereas the heterozygotes are represented by a 1. Homozygotes for the most common allele are represented by a 0.