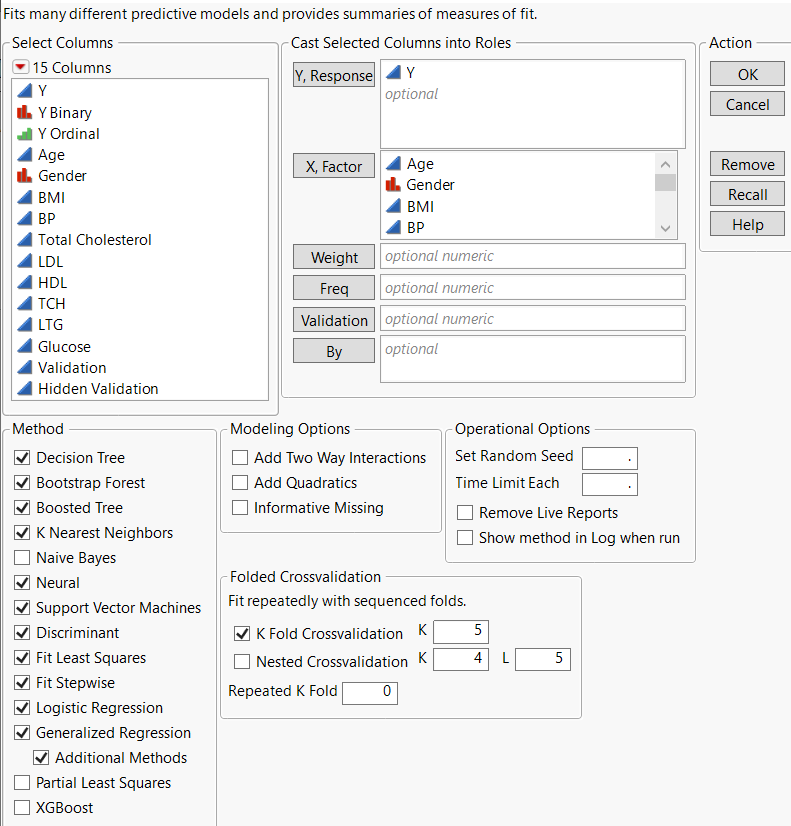
Launch the Model Screening Platform
Launch the Model Screening Platform
To launch the Model Screening platform, select Analyze > Predictive Modeling > Model Screening.
Figure 10.3 The Model Screening Launch Window
For more information about the options in the Select Columns red triangle menu, see “Column Filter Menu” in Using JMP.
Y, Response
The response variable or variables that you want to analyze.
X, Factor
The predictor variables.
Weight
(Not applicable to the K Nearest Neighbors, Support Vector Machines, or Neural modeling platforms.) A column whose numeric values assign a weight to each row in the analysis.
Freq
(Not applicable to the K Nearest Neighbors modeling platform.) A column whose numeric values assign a frequency to each row in the analysis.
Validation
(Not applicable if any of the Crossvalidation options are selected in the launch window.) A numeric column that defines the validation sets. If you click the Validation button with no columns selected in the Select Columns list, you can add a validation column to your data table. For more information about the Make Validation Column utility, see “Make Validation Column”.
Note: If you specify a validation column with more than three levels, this column is used to perform K Fold crossvalidation.
By
A column or columns whose levels define separate analyses. For each level of the specified column, the corresponding rows are analyzed using the other variables that you have specified. The results are presented in separate reports. If more than one By variable is assigned, a separate report is produced for each possible combination of the levels of the By variables.
Methods
Enables you to select the desired modeling platforms. By default, the modeling platforms that are fit are Decision Tree (Partition), Bootstrap Forest, Boosted Tree, K Nearest Neighbors, Neural, Support Vector Machines, Discriminant, Fit Least Squares, Fit Stepwise, Logistic Regression, and Generalized Regression. Naive Bayes, Partial Least Squares, and XGBoost are also available.
Notes:
– XGBoost is not supported by JMP and is available only if the XGBoost add-in is installed. For more information about XGBoost, see community.jmp.com.
– Decision Tree (Partition), Discriminant, and Partial Least Squares all require some type of validation set in order to fit a model.
– If there are fewer than 20 observations in a validation set, a Decision Tree (Partition) model cannot be fit.
– The modeling platforms use default options and tuning parameters in model fitting. You can try to improve the fit past what the default yields by calling platforms directly and choosing different options.
– The Additional Methods option under Generalized Regression calls several additional methods, such as Ridge, Elastic Net and Lasso, in the Generalized Regression platform. For the Lasso method, Early Stopping is disabled when there are less than 1000 observations and less than 100 variables. See “Generalized Regression Models” in Fitting Linear Models.
Caution: This results in additional model fits.
Modeling Options
Provides additional options for the modeling platforms.
Add Two Way Interactions
Adds all two way interaction effects to linear models.
Add Quadratics
Adds effects for the squares of continuous variables to linear models.
Informative Missing
Enables informative missing for all platforms.
Operational Options
Provides additional options.
Set Random Seed
Sets a random seed that is used for any random components of the model fit routines. This enables you to rerun the platform and obtain the same model fits.
Time Limit Each
Specifies a time limit, in seconds, for each fit. For platforms that support early stopping, the best estimates up to that point are provided. For platforms that do not support early stopping, no result is provided.
Remove Live Reports
Does not include the individual model platform reports in the Model Screening report window.
Tip: Select this option to free up memory when you have a large problem with many methods and fits.
Show method in Log when run
Writes out a progress message to the log each time a fitting platform is called.
Folded Crossvalidation
Provides options for various types of crossvalidation.
K Fold Crossvalidation
Divides the data randomly into K parts or folds. A model is fit to the data using K-1 folds to build the model and the remaining fold used for crossvalidation. This is repeated K times for a total of K models. The default value of K is 5.
– K specifies the number of folds for K Fold Crossvalidation. The default is 5 and K must be greater than 1.
– The results for the best model are provided.
Nested Crossvalidation
Divides the data into nested folds for crossvalidation. First, the data are divided into k = 1, ..., K equals parts, or folds. For each fold, the kth fold is used as a test set and the remaining data are divided further into L equal parts. These L subdivisions are called inner folds. Then, a model is fit to the data using L-1 inner folds with the remaining inner fold held out each time as a crossvalidation set. The L models then use the kth fold as a common test data set. In all, a total of K*L models are fit. The default value of K is 4 and the default value of L is 5.
For example, set K = 2 and L = 3. The data are initially divided into two folds. The first fold is held out as a test set and the second fold is divided into 3 inner folds. Three models are fit to the data, each time with a different inner fold held out as a crossvalidation set. Then, all three models are tested on the first fold.
The second fold is then held out as a test set and the first fold is divided into 3 inner folds. Three models are fit to the data, each time with a different inner fold held out as a crossvalidation set. Then, all three models are tested on the second fold.
– K specifies the number of folds for Nested Crossvalidation. The default is 4 and K must be greater than 1.
– L specifies the number of inner folds for Nested Crossvalidation. The default is 5 and L must be greater than 1.
Note: If both K Fold Crossvalidation and Nested Crossvalidation are selected, Nested Crossvalidation is performed.
Repeated K Fold
Specifies the number of times the K Fold Crossvalidation or Nested Crossvalidation process is repeated.
When you click OK, the specified models are fit and a set of progress bars are shown. The upper progress bar reports the progress across all fits. The lower progress bar reports the progress for the current individual model fit. You can stop the lower progress bar to use early stopping and the upper progress bar will continue to run.