Example of a Split Plot Experiment
Example of a Split Plot Experiment
In this example, use the Mixed Model personality in the Fit Model platform to specify and analyze a split-plot experiment with two factors. When you specify the Mixed Model personality, the Fit Model launch window provides tabs for specifying effects. The resulting analysis is targeted to random effects. Note, however, that split-plot experiments can also be analyzed using the Standard Least Squares personality.
The data in this example come from a study of the effects of tenderizer and length of cooking time on meat. Six beef carcasses were randomly selected from carcasses at a meat packaging plant. From the right rib-eye muscle of each carcass, three rolled roasts were prepared under uniform conditions. Each of these three roasts was assigned a tenderizing treatment at random. After treatment, a coring device was used to mark four cores of meat near the center of each.
The three roasts from the same carcass were placed together in a preheated oven and allowed to cook. After 30 minutes, one of the cores was taken at random from each roast. Cores were removed in this fashion again after 36 minutes, 42 minutes, and 48 minutes. As each set cooled to serving temperature, the cores were measured for tenderness using the Warner-Bratzler device. Larger measurements indicate tougher meat.
Your interest centers on the effects of tenderizer, roasting time, and especially whether there is an interaction between tenderizer and roasting time. This design addresses that goal.
1. Select Help > Sample Data Folder and open Split Plot.jmp.
2. Select Analyze > Fit Model.
3. Select Y and click Y.
4. Select Mixed Model from the Personality list.
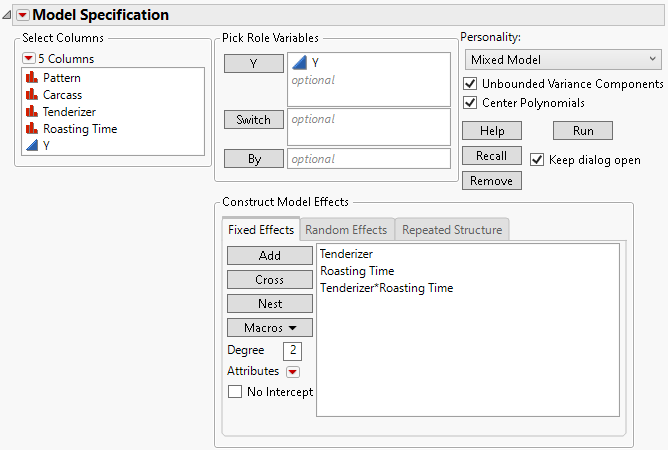
5. Select Tenderizer and Roasting Time, and then select Macros > Full Factorial.
Figure 8.27 Fit Model Launch Window Showing Completed Fixed Effects Tab
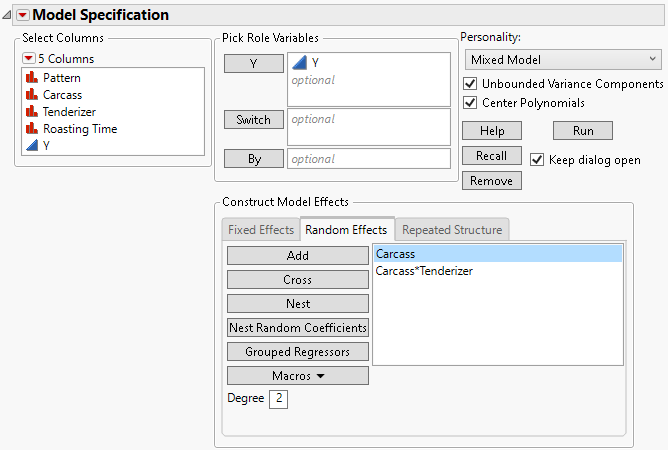
6. Select the Random Effects tab.
7. Select Carcass and click Add to create the random carcass effect.
8. Select Carcass and Tenderizer and click Cross.
The Carcass*Tenderizer interaction is the error term for the whole plot factor, Tenderizer. This is equivalent to the Carcass*Tenderizer&Random term in Standard Least Squares.
Figure 8.28 Fit Model Launch Window Showing Completed Random Effects Tab
9. Click Run.
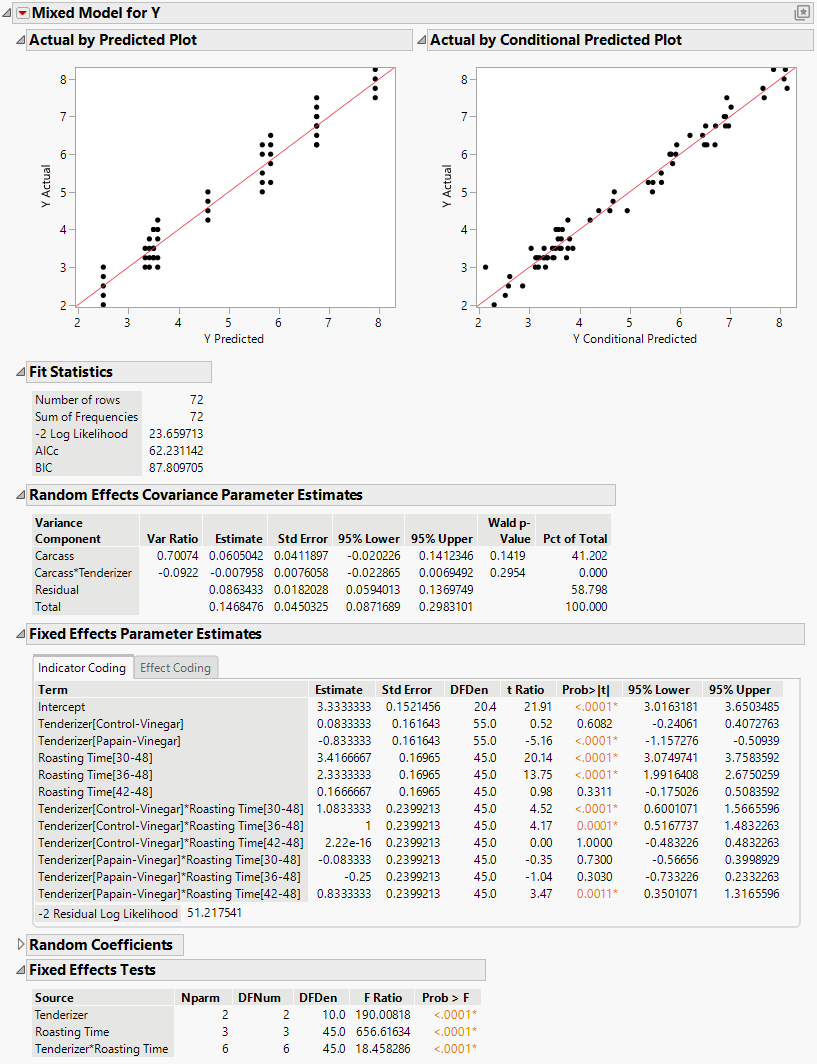
The Mixed Model report is shown in Figure 8.29.
The Actual by Predicted Plot and the Actual by Conditional Predicted Plot show no issues with model fit, so you can proceed to interpret the results. The Fixed Effects Tests report indicates that there is a significant interaction between tenderizer and roasting time.
Figure 8.29 Mixed Model Report
Explore the Interaction between Tenderizer and Roasting Time
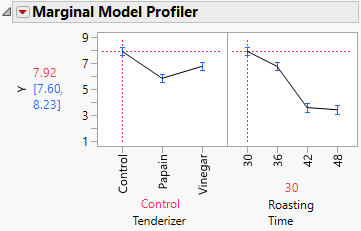
1. Click the Mixed Model red triangle and select Marginal Model Inference > Profiler.
Figure 8.30 Marginal Model Profiler with Roasting Time Set to 30 Minutes
2. Move the red dashed vertical line in the Roasting Time panel to 36, 42, and 48.
In Figure 8.30, notice that both the papain and vinegar tenderizers result in significantly lower tenderness scores than the control when roasting time is either 30 or 36 minutes. However, at 42 minutes, there are no significant differences. At 48 minutes, papain gives a value lower than the control, but vinegar does not. Papain gives lower tenderness scores than does vinegar at all times except 42 minutes.
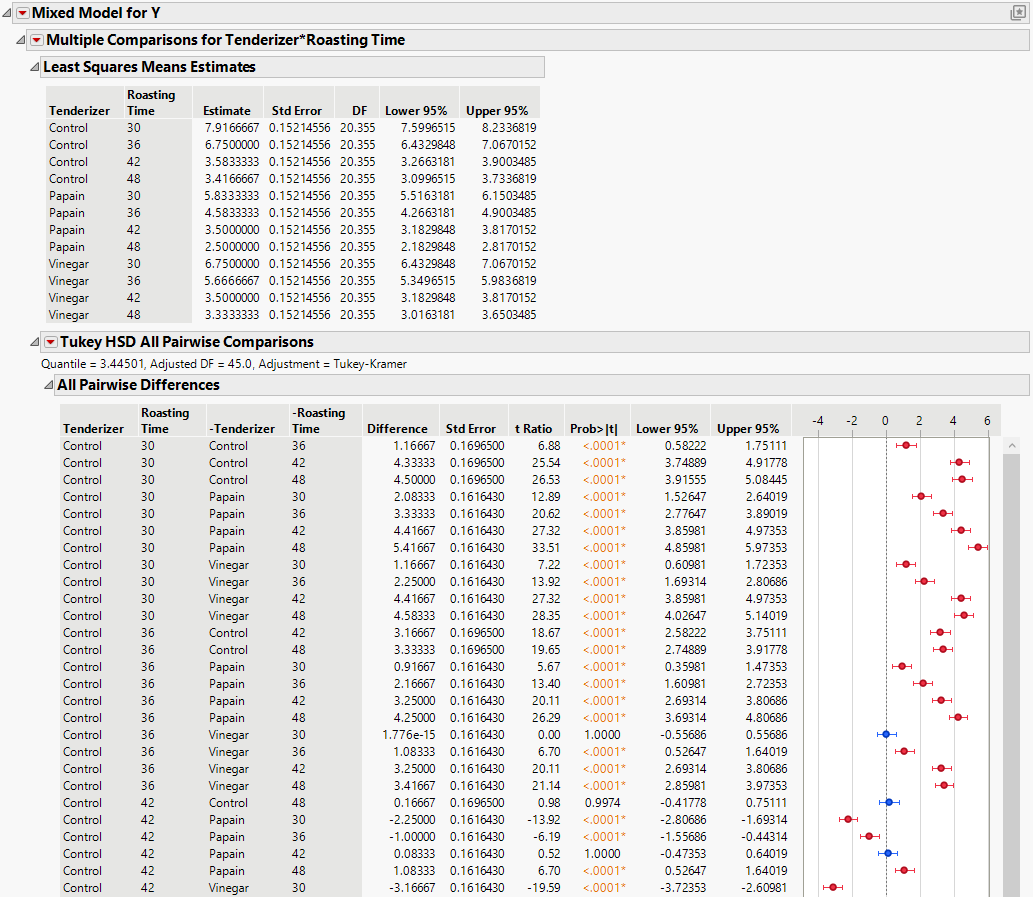
3. Click the Mixed Model red triangle and select Multiple Comparisons.
4. Select Tenderizer*Roasting Time.
5. Select All Pairwise Comparisons - Tukey HSD and then click OK.
Figure 8.31 shows a partial list of pairwise comparisons. Most of the differences between papain and vinegar that you observed in the profiler are statistically significant. Therefore, it appears that papain is the better tenderizer.
Figure 8.31 Multiple Comparisons, Partial View