Process Description
Ingenuity Get Gene Pathways
The Ingenuity Get Gene Pathways process retrieves all of the pathway identifiers for all of the genes in the input data set and creates a new column containing them. The new column includes the delimiter " / " to separate the pathway identifiers, and is suitable for use with the Gene Set Enrichment and Gene Set Scoring processes, or for general searching.
Note: This process is considered experimental.
What do I need?
Two files are required to run the Ingenuity Get Gene Pathways process:
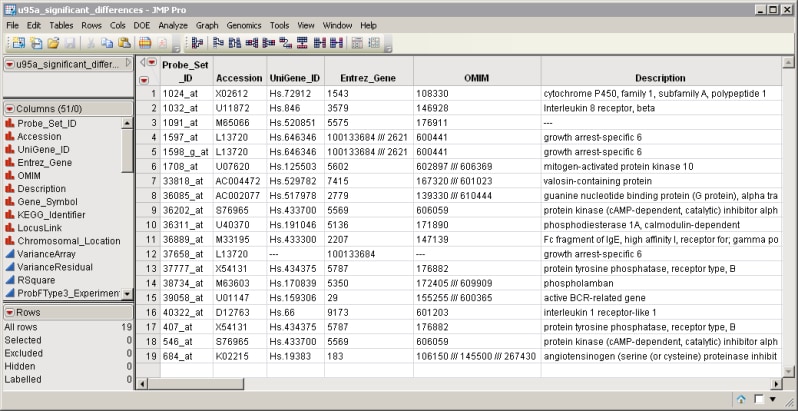
| • | A SAS Input Data Set that must contain a variable with valid Entrez gene identifiers. The u95a_significant_differences.sas7bdat data set serves as an example, and is shown below. This data set contains numerous variables listing various gene identifiers. |
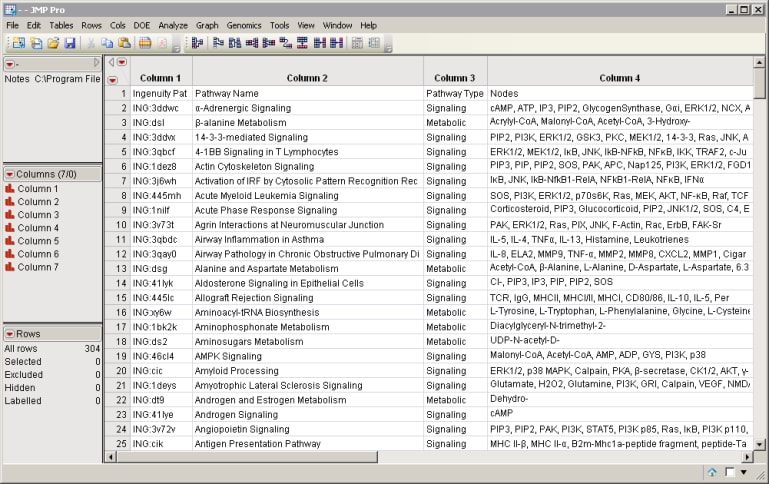
| • | The Ingenuity Canonical Pathway Membership Data file. This CSV file, which can be downloaded from Ingenuity, lists the identifiers, nodes, and associated genes for each pathway in the Ingenuity database. |
Note: If you are an IPA user who needs help with downloading the Ingenuity pathway list, contact Ingenuity customer support by e-mail at support@ingenuity.com.
The u95a_significant_differences.sas7bdat data set is located in the Sample Data\Microarray\Affymetrix Latin Square directory included with JMP Genomics. The IngenuityIdList.xls Ingenuity pathways file was downloaded from Ingenuity and placed in the C:\Program Files\SASHome\JMPGenomics\13\Genomics\Testing\ThirdPartyAnnotation folder created for this example.
For detailed information about the files and data sets used or created by JMP Genomics software, see Files and Data Sets.
Output/Results
Refer to the Ingenuity Get Gene Pathways output documentation for detailed descriptions of the output of this process.