Process Description
Bivariate One-Way ANOVA
Bivariate One-Way ANOVA fits a one-way repeated-measures ANOVA linear model to pairs of rows of the Input Data Set. This model can be applied to allele-level intensity data to simultaneously test for both copy number and allele ratio changes.
For hypothesis testing, two sets of Volcano Plots are generated. The first set contains one-way mean differences that are averaged across the bivariate observations. The second set contains one-way mean differences that are differenced across the bivariate observations. For bivariate allele intensity data, the first set tests for copy number changes and second set tests for allele frequency changes. Significant results from the second set could be caused by loss of heterozygosity (LOH).
Note: This process can be computationally intensive for large data sets, but runs faster than Mixed Model Analysis for identical models.
What do I need?
Two data sets are required for this process:
| • | The Input Data Set that contains all of the numeric data to be analyzed. The data should be normalized, so that global effects (such as dye, chip to chip variation, and so on) are removed. Data should be on the log2 scale. The data values are responses; for example, intensity measurements, and are assumed to be independent with Gaussian errors. The data set must be in the tall format, in which columns are arrays or samples, and rows are molecular entities, such as SNPs or probesets. Furthermore, pairs of rows should be associated with each other (for example, having the same probeset identifier). An example is shown below. |
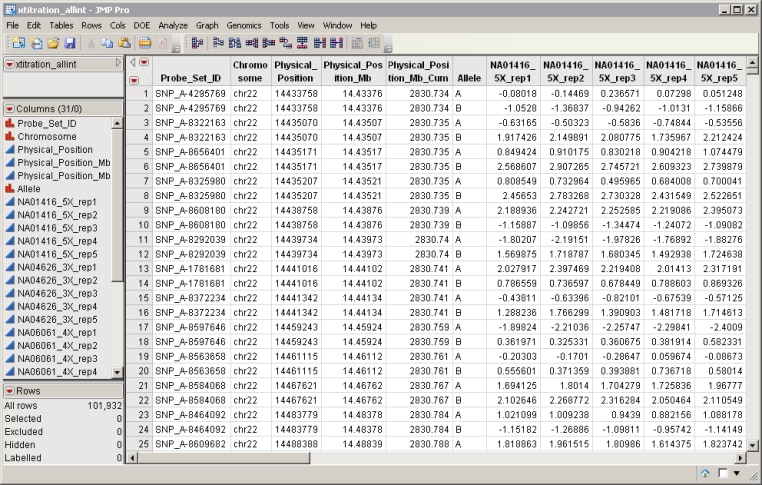
Note: The bivariate observations are assumed to have the same variance, and a correlation is modeled between them. If the mean or covariance structure is more complex than this, then you should run the Mixed Model Analysis processes instead.
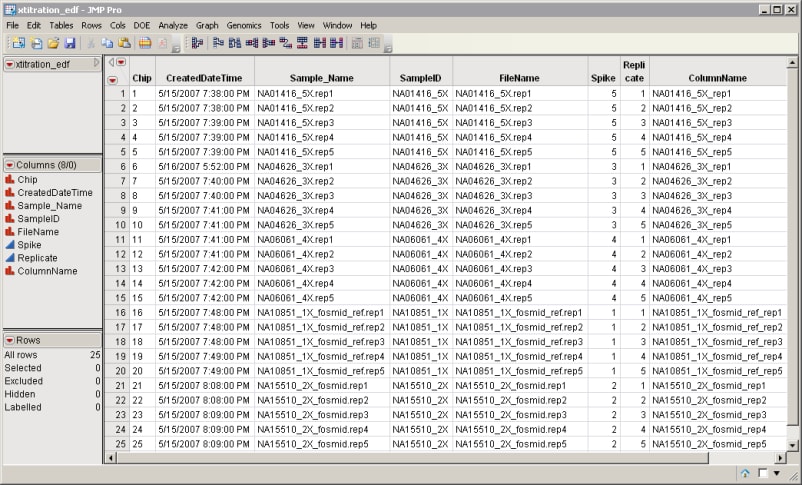
| • | The Experimental Design Data Set (EDDS). This data set tells how the experiment was performed, providing information about the columns in the input data set. Note that one column in the EDDS must be named ColumnName. The purpose of this column is to index the names of the columns in the input data set; values contained in this column must exactly match the column names in the input data set. Two other columns in this data set, Array and Experiment, correspond to an index variable and the experimental variable, respectively. An example of an EDDS is shown below. |
An Annotation Data Set is optional. This data set contains information, such as gene identity or chromosomal location, for each of the rows in the input data set. This data set is also in the tall format; where each row corresponds to a different molecular entity. The variable that identifies those entities (in this case Probe_Set_ID) is normally used as a merge key to join the input data set to the annotation data.
For detailed information about the files and data sets used or created by JMP Genomics software, see Files and Data Sets.
Output/Results
The output generated by this process is summarized in a Tabbed report. Refer to the Bivariate One-Way ANOVA output documentation for detailed descriptions and guides to interpreting your results.