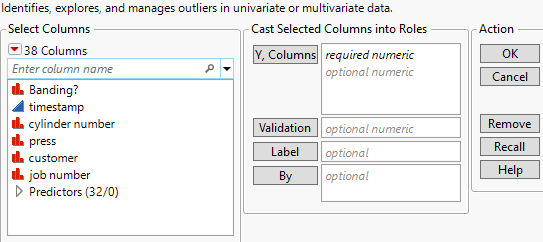
Launch the Explore Outliers Platform
To launch Explore Outliers, select Analyze > Screening > Explore Outliers.
Note: The Explore Outliers commands analyze only columns with a Continuous modeling type. Other columns can be entered in the launch window but are ignored.
Figure 21.5 Explore Outliers Platform Launch Window
For more information about the options in the Select Columns red triangle menu, see “Column Filter Menu” in Using JMP.
Y, Columns
Specifies the columns that you want to analyze.
Validation
Specifies a validation column that is used for Robust PCA Outliers.
Label
Specifies a column that replaces row numbers in multivariate analysis reports with labels.
By
A column or columns whose levels define separate analyses. For each level of the specified column, the corresponding rows are analyzed using the other variables that you have specified. The results are presented in separate reports. If more than one By variable is assigned, a separate report is produced for each possible combination of the levels of the By variables.
Tip: To run an outlier analysis across all levels of a By variable, press Ctrl and click the desired outlier analysis command button.
After you click OK, the Explore Outliers report appears. The report contains several methods for finding outliers in univariate and multivariate data. There are options for each method that you can specify before making a selection.
Univariate
There are two options for exploring outliers in your univariate data.
Quantile Range Outliers
Uses the quantile distribution of each column to identify outliers as extreme values. This tool is useful for discovering missing value or error codes within the data. This is the recommended method to begin exploring outliers in your data. See Quantile Range Outliers. You can specify the following options:
Tail Quantile
The probability for the lower quantile that is used to calculate the interquantile range. The probability of the upper quantile is considered 1 - Tail Quantile. For example, a Tail Quantile value of 0.1 means that the interquantile range is between the 0.1 and 0.9 quantiles of the data. The default value is 0.1.
Q
The multiplier that determines the outlier threshold. Values that fall beyond Q times the interquantile range past the Tail Quantile or 1 - Tail Quantile values are identified as outliers. Large values of Q provide a more conservative set of outliers than small values. The default value is 3.
Robust Fit Outliers
Finds robust estimates of the center and spread of each column and identifies outliers as those data points that are far from those values. See Robust Fit Outliers. You can specify the following options:
K Sigma
The multiplier that determines outliers as K times the spread away from the center. Large values of K provide a more conservative set of outliers than small values. The default value is 4.
Huber
Uses Huber M-Estimation to estimate center and spread. This option is the default. See Huber and Ronchetti (2009).
Cauchy
Assumes a Cauchy distribution to calculate estimates for the center and spread. Cauchy estimates have a high breakdown point and are typically more robust than Huber estimates. However, if your data are separated into clusters, the Cauchy distribution tends to consider only the half of the data that is clustered more closely, ignoring the rest.
Quartile
Uses the median as the measure of center and the interquartile range (IQR) divided by 1.34898 as the measure of spread. Dividing the IQR by the 1.34898 factor results in the spread corresponding to one standard deviation if the data are normally distributed.
Multivariate
There are two options for exploring outliers in your multivariate data.
Robust PCA Outliers
Decomposes data into a low-rank matrix and residuals and uses the residuals to detect outliers. See Robust PCA Outliers. You can specify a value for Lambda and select if the data should be centered. For advanced options, access the Robust PCA Outliers options window by pressing Shift and clicking the Robust PCA Outliers button.
Lambda
Specifies a value that determines the sparsity of the matrix of residuals. For larger values of Lambda, the matrix of residuals is more sparse. For a data table with n training rows and p columns, the default value of Lambda is defined as follows:
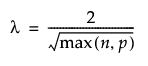
Max Iterations
Specifies the maximum number of SVD iterations. The default number of iterations is 100. If there are more than 20,000 columns specified in the launch, the default number of iterations is 50.
Note: If the algorithm does not converge after the maximum number of iterations, a JMP alert is shown. You can continue with more iterations or cancel. If you click Cancel and a less stringent convergence criterion is met, the results are shown. If you click Cancel and the less stringent convergence criterion is not met, another JMP alert is shown to ask whether or not to accept the results.
Converge Criterion
Determines when to stop the algorithm. The default convergence criterion values are set based on the number of columns specified in the launch.
• If the number of columns is less than 2,000, the default is 1e-7.
• If the number of columns is greater than or equal to 2,000, the default is 1e-6.
• If the number of columns is greater than or equal to 20,000, the default is 1e-5.
• The less stringent convergence criterion is set to1000 times the original convergence criterion.
Outlier Threshold
Specifies the outlier threshold that determines which outliers are shown in the Cell Large Residuals table. An observation is shown if the scaled residuals is larger than the following:
min[0.99 × max{abs(scaled residuals)}, Outlier Threshold]
By default, the Outlier Threshold is 2. If using 2 as the Outlier Threshold results in more than a million outliers, the Outlier Threshold is changed to 3.
Center
Determines whether the data are centered before the Robust PCA Outlier algorithm is performed.
Note: If the number of rows is less than or equal to 10, the data are not centered.
Scale
Determines whether the data are scaled before the Robust PCA Outlier algorithm is performed.
Note: If the number of rows is less than or equal to 10, the data are not scaled.
Randomized SVD for Very Wide Problems
(Available only if the number of columns specified in the launch is greater than or equal to 1000.) Uses the Randomized SVD method instead of the Lanczos method to decompose the data. This option speeds up the Robust PCA Outlier calculations for wide data. See “Randomized SVD”.
Randomized Dimensions
(Available only if the number of columns specified in the launch is greater than or equal to 1000.) Specifies the number of dimensions used in Randomized SVD.
K Nearest Neighbor Outliers
Identifies outliers as values that are far from their k-nearest neighbors. See K Nearest Neighbor Outliers. You can specify the following options:
K
Specifies the upper bound for the farthest neighbor to be considered. The default value is 8.
Impute Missing
Specifies whether missing values are imputed. If selected, missing values are imputed using Multivariate RPCA Imputation. See “Multivariate RPCA Imputation”.