Output Description
Quantitative TDT
Running this process using the GeneticMarkerExample sample setting generates the tabbed Results window shown below. Refer to the Quantitative TDT process description for more information. Output from the process is organized into tabs. Each tab contains one or more plots, data panels, data filters, and so on. that facilitate your analysis.
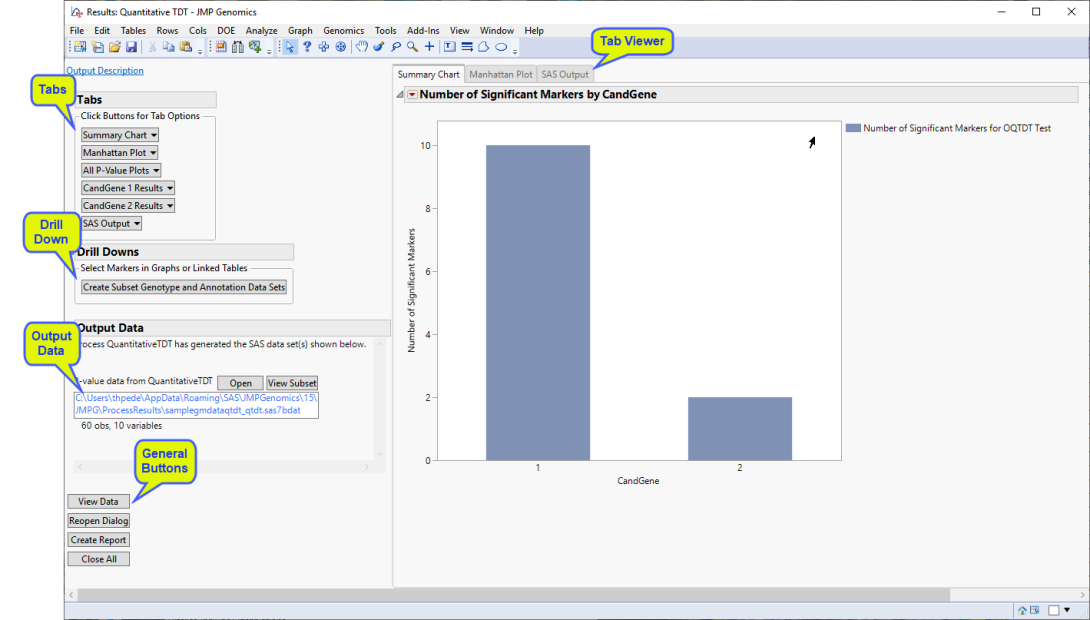
The Results window contains the following panes:
Tabs
This pane enables you to access and view the output plots and associated data sets on each tab. Use the drop-down menu to view the tab in the Tab Viewer pane, open the tab in a new window, or remove the tab and its contents from the Tab Viewer pane.
TThe following tabs are generated by this process:
| • | Summary Chart: When there are multiple annotation groups (chromosomes or genes, for example), this tab displays the number of significant markers in each annotation group for each test. Separate bar charts are shown for each BY group when any BY variables are specified. This tab is open by default. |
| • | Manhattan Plot: When there are multiple annotation groups (chromosomes or genes, for example), this tab displays a scatter plot of the p-values across all annotation groups. |
| • | Annotation Group Results: When there are multiple annotation groups (chromosomes or genes, for example), a separate Results tab with an overlay plot of p-value by chromosome location is created for each annotation group. If the Calculate trend odds ratios check box was checked, this tab also contains a Volcano Plot of p-value by log odds ratio for all markers. |
In this example, there are two annotation groups (CandGene 1, and CandGene 2) and, thus two Annotation Group Results tabs (CandGene 1 Results and CandGene 2 Results).
| • | All P-Value Plots: When there are multiple annotation groups (chromosomes or genes, for example), the All P-Value Plots tab shows all the p-value plots from the Annotation Group Results tabs in a single display. |
Note: When an annotation group variable is not specified or there is only one annotation group, the tab is named P-Value Plot and contains an overlay plot of p-value by chromosome location for all markers.
| • | SAS Output: This tab displays the SAS output from the SAS procedures used to perform the quantitative TDT. |
Drill Downs
Action buttons provide you with an easy way to drill down into your data. The following action buttons are generated by this process:
| • | Create Subset Genotype and Annotation Data Sets: Select points from the p-value plots and click to open the Subset and Reorder Genetic Data process to create the subset data sets. |
Note: This action button is not available if any By Variables are selected.
Output Data
This process generates the following data set(s):
| • | P-value Data Set: This data set contains all the columns from the annotation data set, plus the test statistics and p-values from the tests performed. This data set can be used as the annotation data set for subsequent processes to accumulate results from multiple processes into a single data set. The name of this data is set is given by the Output File Prefix, or input data set name if none given, with the suffix _qtdt. Click to view the data set. |
For detailed information about the files and data sets used or created by JMP Genomics software, see Files and Data Sets.
Tab Viewer
This pane provides you with a space to view individual tabs within the Results window.
General
| • | Click to reveal the underlying data table associated with the current tab. |
| • | Click to reopen the completed process dialog used to generate this output. |
| • | Click to generate a pdf- or rtf-formatted report containing the plots and charts of selected tabs. |
| • | Click to close all graphics windows and underlying data sets associated with the output. |